Omics-analysis
Seeding comprehensive analysis in their named directories (e.g., BMI), the repository links to technical issues documented in physalia, Mixed-Models, software-notes and other sister repositories: SUMSTATS, FM-pipeline, PW-pipeline, hess-pipeline, TWAS-pipeline, EWAS-fusion. for fine-mapping, pathway analysis, TWAS, Mendelian randomisation, predictive analytics and other topics as highlighted in the wiki page.
Earlier or broader aspects have been reflected in the following repositories: Haplotype-Analysis, misc, R.
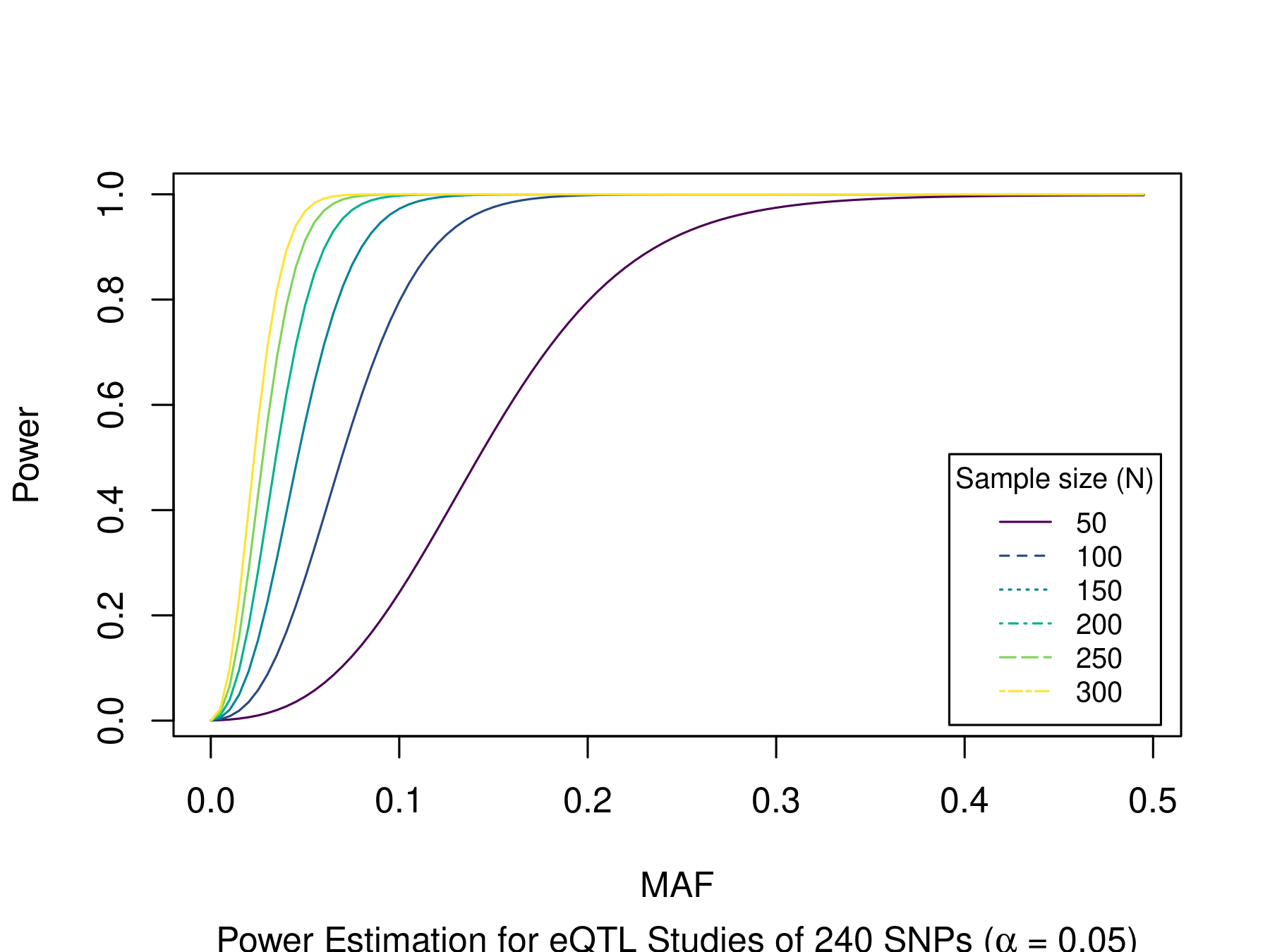
The figure below was generated with eQTL.R.