This package intends to gather information, metadata and relevant scripts in proteogenomic analysis.
1 Collections
As used in several years of proteomic analysis and for future extensions, the collections are in two locations:
-
data/. R datasets. -
inst/.EndNote/,Olink/,scripts/,SomaLogic/, which spread into the package’s root directory after installation.
While library(help=pQTLdata) displays the general information, ? pQTLdata can give a list of data objects in the package.
2 Protein panels
2.1 Caprion
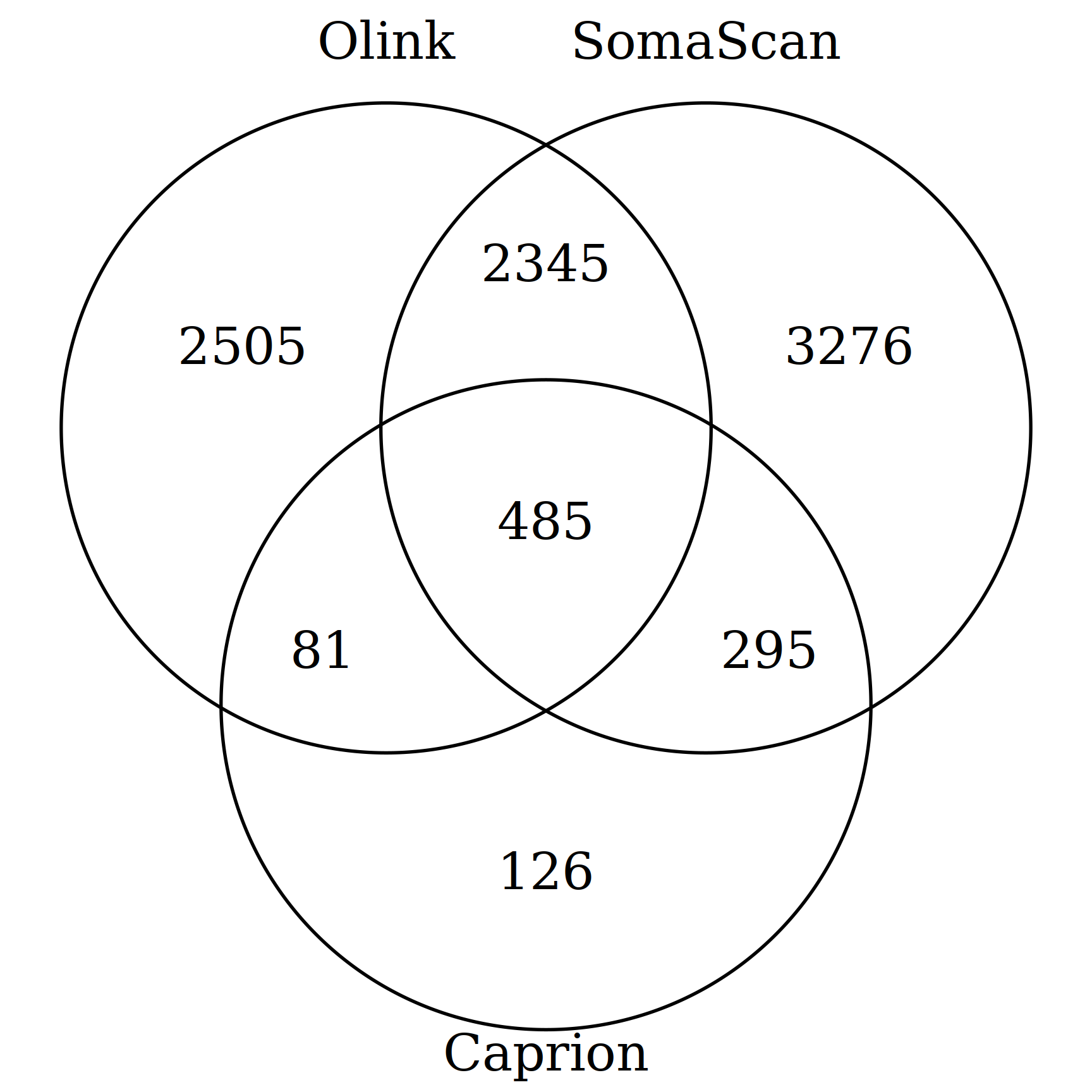
As has been the norm, no snapshot upon data release was provided which consequently requires substantial effort and the notable ones are highlighted here.
| Gene | Gene.orig | Protein | Accession | ensGenes | chr | start | end |
|---|---|---|---|---|---|---|---|
| AMY1 | AMY1A; AMY1B; AMY1C | AMY1_HUMAN | P04745 | ENSG00000174876, ENSG00000187733, ENSG00000237763 | 1 | 103655760 | 103758726 |
| C20orf27 | C20orf27 | CT027_HUMAN | Q9GZN8 | ENSG00000101220 | 20 | 3753508 | 3767781 |
| C4B | C4B; C4B_2 | CO4B_HUMAN | P0C0L5 | ENSG00000224389, ENSG00000236625, ENSG00000228267, ENSG00000233312, ENSG00000228454 | 6 | 3262537 | 32035418 |
| C7orf26 | C7orf26 | CG026_HUMAN | Q96N11 | ENSG00000146576 | 7 | 6590021 | 6608726 |
| CD97 | CD97 | CD97_HUMAN | P48960 | ENSG00000123146 | 19 | 14380501 | 14408725 |
| FAM49B | FAM49B | FA49B_HUMAN | Q9NUQ9 | ENSG00000153310 | 8 | 129839593 | 130017504 |
| GBA | GBA | GLCM_HUMAN | P04062 | ENSG00000177628 | 1 | 155234452 | 155244699 |
| HBA | HBA1; HBA2 | HBA_HUMAN | P69905 | ENSG00000188536, ENSG00000206172 | 16 | 172876 | 177522 |
| HIST | HIST1H4A; HIST1H4B; HIST1H4C; HIST1H4D; HIST1H4E; HIST1H4F; HIST1H4H; HIST1H4I; HIST1H4J; HIST1H4K; HIST1H4L; HIST2H4A; HIST2H4B; HIST4H4 | H4_HUMAN | P62805 | ENSG00000277157, ENSG00000275126, ENSG00000274618, ENSG00000197061, ENSG00000158406, ENSG00000197238, ENSG00000276966, ENSG00000197837, ENSG00000276180, ENSG00000278705, ENSG00000273542, ENSG00000278637, ENSG00000270276, ENSG00000270882 | 6, 12, 1 | 14767999 | 149861159 |
| SLC9A3R1 | SLC9A3R1 | NHRF1_HUMAN | O14745 | ENSG00000109062 | 17 | 74748628 | 74769353 |
| WARS | WARS | SYWC_HUMAN | P23381 | ENSG00000140105 | 14 | 100333790 | 100376805 |
| YARS | YARS | SYYC_HUMAN | P54577 | ENSG00000134684 | 1 | 32775237 | 32818031 |
which again is useful for extracting data from GTEx v8.
3 Published data
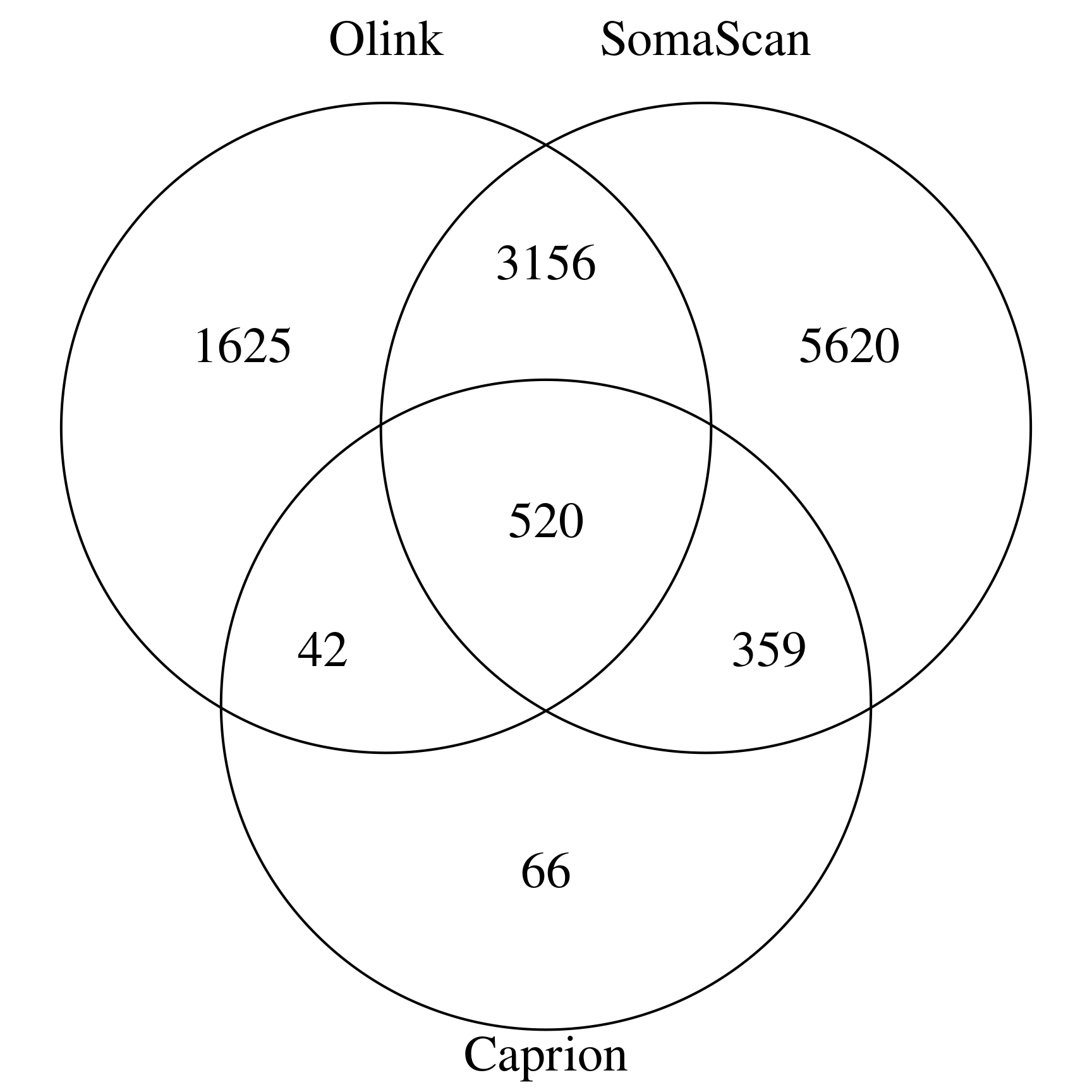
This associates with several panels, including SomaScan (SomaScan160410)1, Olink qPCR inflammation
(inf1)2 and Seer (seer1980)3. In particular, inf1 contains updates from the original release by Olink.
| gene | prot | uniprot | target | target.short | chr | start | end | ensembl_gene_id | alt_name | ensGene | chromosome | start38 | end38 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ADA | ADA | P00813 | Adenosine Deaminase (ADA) | ADA | 20 | 43248163 | 43280874 | ENSG00000196839 | NA | NA | NA | NA | NA |
| ARTN | ARTN | Q5T4W7 | Artemin (ARTN) | ARTN | 1 | 44398992 | 44402913 | ENSG00000117407 | NA | NA | NA | NA | NA |
| AXIN1 | AXIN1 | O15169 | Axin-1 (AXIN1) | AXIN1 | 16 | 337440 | 402673 | ENSG00000103126 | NA | NA | NA | NA | NA |
| BDNF | BDNF | P23560 | Brain-derived neutrophic factor (BDNF) | BDNF | 11 | 27676440 | 27743605 | ENSG00000176697 | NA | NA | NA | NA | NA |
| CASP8 | CASP.8 | Q14790 | Caspase 8 (CASP-8) | CASP-8 | 2 | 202098166 | 202152434 | ENSG00000064012 | NA | NA | NA | NA | NA |
| CCL11 | CCL11 | P51671 | Eotaxin-1 (CCL11) | CCL11 | 17 | 32612687 | 32615353 | ENSG00000172156 | CCL11 | NA | NA | NA | NA |
| CCL13 | MCP.4 | Q99616 | Monocyte chemotactic protein 4 (MCP-4) | MCP-4 | 17 | 32683471 | 32685629 | ENSG00000181374 | CCL13 | NA | NA | NA | NA |
| CCL19 | CCL19 | Q99731 | C-C motif chemokine 19 (CCL19) | CCL19 | 9 | 34689564 | 34691274 | ENSG00000172724 | NA | NA | NA | NA | NA |
| CCL2 | MCP.1 | P13500 | Monocyte chemotactic protein 1 (MCP-1) | MCP-1 | 17 | 32582304 | 32584222 | ENSG00000108691 | CCL2 | NA | NA | NA | NA |
| CCL20 | CCL20 | P78556 | C-C motif chemokine 20 (CCL20) | CCL20 | 2 | 228678558 | 228682272 | ENSG00000115009 | NA | NA | NA | NA | NA |
| CCL23 | CCL23 | P55773 | C-C motif chemokine 23 (CCL23) | CCL23 | 17 | 34340096 | 34345005 | ENSG00000167236 | NA | ENSG00000274736 | 17 | 36013056 | 36017972 |
| CCL25 | CCL25 | O15444 | C-C motif chemokine 25 (CCL25) | CCL25 | 19 | 8117651 | 8127534 | ENSG00000131142 | NA | ENSG00000131142 | 19 | 8052318 | 8062660 |
| CCL28 | CCL28 | Q9NRJ3 | C-C motif chemokine 28 (CCL28) | CCL28 | 5 | 43376747 | 43412493 | ENSG00000151882 | NA | NA | NA | NA | NA |
| CCL3 | MIP.1.alpha | P10147 | Macrophage inflammatory protein 1-alpha (MIP-1 alpha) | MIP-1 alpha | 17 | 34415602 | 34417515 | ENSG00000006075 | NA | ENSG00000277632 | 17 | 36088256 | 36090169 |
| CCL4 | CCL4 | P13236 | C-C motif chemokine 4 (CCL4) | CCL4 | 17 | 34430983 | 34433014 | ENSG00000129277 | NA | ENSG00000275302 | 17 | 36103827 | 36105621 |
| CCL7 | MCP.3 | P80098 | Monocyte chemotactic protein 3 (MCP-3) | MCP-3 | 17 | 32597240 | 32599261 | ENSG00000108688 | CCL7 | NA | NA | NA | NA |
| CCL8 | MCP.2 | P80075 | Monocyte chemotactic protein 2 (MCP-2) | MCP-2 | 17 | 32646055 | 32648421 | ENSG00000108700 | CCL8 | NA | NA | NA | NA |
| CD244 | CD244 | Q9BZW8 | Natural killer cell receptor 2B4 (CD244) | CD244 | 1 | 160799950 | 160832692 | ENSG00000122223 | NA | NA | NA | NA | NA |
| CD274 | PD.L1 | Q9NZQ7 | Programmed cell death 1 ligand 1 (PD-L1) | PD-L1 | 9 | 5450503 | 5470566 | ENSG00000120217 | NA | NA | NA | NA | NA |
| CD40 | CD40 | P25942 | CD40L receptor (CD40) | CD40 | 20 | 44746911 | 44758502 | ENSG00000101017 | NA | NA | NA | NA | NA |
| CD5 | CD5 | P06127 | T-cell surface glycoprotein CD5 (CD5) | CD5 | 11 | 60869867 | 60895324 | ENSG00000110448 | NA | NA | NA | NA | NA |
| CD6 | CD6 | P30203 | T-cell surface glycoprotein CD6 isoform (CD6) | CD6 | 11 | 60739115 | 60787849 | ENSG00000138675 | NA | ENSG00000013725 | 11 | 60971680 | 61020377 |
| CDCP1 | CDCP1 | Q9H5V8 | CUB domain-containing protein 1 (CDCP1) | CDCP1 | 3 | 45123770 | 45187914 | ENSG00000163814 | NA | NA | NA | NA | NA |
| CSF1 | CSF.1 | P09603 | Macrophage colony-stimulating factor 1 (CSF-1) | CSF-1 | 1 | 110452864 | 110473614 | ENSG00000184371 | NA | NA | NA | NA | NA |
| CST5 | CST5 | P28325 | Cystatin D (CST5) | CST5 | 20 | 23856572 | 23860387 | ENSG00000170367 | NA | NA | NA | NA | NA |
| CX3CL1 | CX3CL1 | P78423 | Fractalkine (CX3CL1) | CX3CL1 | 16 | 57406370 | 57418960 | ENSG00000006210 | NA | NA | NA | NA | NA |
| CXCL1 | CXCL1 | P09341 | C-X-C motif chemokine 1 (CXCL1) | CXCL1 | 4 | 74735110 | 74736959 | ENSG00000163739 | NA | NA | NA | NA | NA |
| CXCL10 | CXCL10 | P02778 | C-X-C motif chemokine 10 (CXCL10) | CXCL10 | 4 | 76942273 | 76944650 | ENSG00000169245 | NA | NA | NA | NA | NA |
| CXCL11 | CXCL11 | O14625 | C-X-C motif chemokine 11 (CXCL11) | CXCL11 | 4 | 76954835 | 76962568 | ENSG00000169248 | NA | NA | NA | NA | NA |
| CXCL5 | CXCL5 | P42830 | C-X-C motif chemokine 5 (CXCL5) | CXCL5 | 4 | 74861359 | 74864496 | ENSG00000163735 | NA | NA | NA | NA | NA |
| CXCL6 | CXCL6 | P80162 | C-X-C motif chemokine 6 (CXCL6) | CXCL6 | 4 | 74702214 | 74714781 | ENSG00000124875 | CXCL6 | NA | NA | NA | NA |
| CXCL9 | CXCL9 | Q07325 | C-X-C motif chemokine 9 (CXCL9) | CXCL9 | 4 | 76922428 | 76928641 | ENSG00000138755 | NA | NA | NA | NA | NA |
| DNER | DNER | Q8NFT8 | Delta and Notch-like epidermal growth factor related receptor (DNER) | DNER | 2 | 230222345 | 230579274 | ENSG00000187957 | NA | NA | NA | NA | NA |
| EIF4EBP1 | 4E.BP1 | Q13541 | Eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) | 4EBP1 | 8 | 37887859 | 37917883 | ENSG00000187840 | NA | NA | NA | NA | NA |
| FGF19 | FGF.19 | O95750 | Fibroblast growth factor 19 (FGF-19) | FGF-19 | 11 | 69513000 | 69519410 | ENSG00000162344 | NA | NA | NA | NA | NA |
| FGF21 | FGF.21 | Q9NSA1 | Fibroblast growth factor 21 (FGF-21) | FGF-21 | 19 | 49258816 | 49261587 | ENSG00000105550 | NA | NA | NA | NA | NA |
| FGF23 | FGF.23 | Q9GZV9 | Fibroblast growth factor 23 (FGF-23) | FGF-23 | 12 | 4477393 | 4488894 | ENSG00000118972 | NA | NA | NA | NA | NA |
| FGF5 | FGF.5 | P12034 | Fibroblast growth factor 5 (FGF-5) | FGF-5 | 4 | 81187753 | 81257834 | ENSG00000013725 | NA | ENSG00000138675 | 4 | 80266639 | 80336680 |
| FLT3LG | Flt3L | P49771 | Fms-related tyrosine kinase 3 ligand (FIt3L) | FIt3L | 19 | 49977464 | 49989488 | ENSG00000090554 | NA | NA | NA | NA | NA |
| GDNF | GDNF | P39905 | Glial cell line-derived neutrophic factor (hGDNF) | hGDNF | 5 | 37812779 | 37839788 | ENSG00000168621 | NA | NA | NA | NA | NA |
| HGF | HGF | P14210 | Hepatocyte growth factor (HGF) | HGF | 7 | 81328322 | 81399754 | ENSG00000019991 | NA | NA | NA | NA | NA |
| IFNG | IFN.gamma | P01579 | Interferon gamma (IFN-gamma) | IFN-gamma | 12 | 68548548 | 68553527 | ENSG00000111537 | NA | NA | NA | NA | NA |
| IL10 | IL.10 | P22301 | Interleukin-10 (IL-10) | IL-10 | 1 | 206940947 | 206945839 | ENSG00000136634 | NA | NA | NA | NA | NA |
| IL10RA | IL.10RA | Q13651 | Interleukin-10 receptor subunit alpha (IL-10RA) | IL-10RA | 11 | 117857063 | 117872196 | ENSG00000110324 | NA | NA | NA | NA | NA |
| IL10RB | IL.10RB | Q08334 | Interleukin-10 receptor subunit beta (IL10RB) | IL10RB | 21 | 34638663 | 34669539 | ENSG00000243646 | NA | NA | NA | NA | NA |
| IL12B | IL.12B | P29460 | Interleukin-12 subunit beta (IL-12B) | IL-12B | 5 | 158741791 | 158757895 | ENSG00000113302 | NA | NA | NA | NA | NA |
| IL13 | IL.13 | P35225 | Interleukin-13 (IL-13) | IL-13 | 5 | 131991955 | 131996802 | ENSG00000169194 | NA | NA | NA | NA | NA |
| IL15RA | IL.15RA | Q13261 | Interleukin-15 receptor subunit alpha (IL-15RA) | IL-15RA | 10 | 5990855 | 6020150 | ENSG00000134470 | NA | NA | NA | NA | NA |
| IL17A | IL.17A | Q16552 | Interleukin-17A (IL-17A) | IL-17A | 6 | 52051185 | 52055436 | ENSG00000112115 | NA | NA | NA | NA | NA |
| IL17C | IL.17C | Q9P0M4 | Interleukin-17C (IL-17C) | IL-17C | 16 | 88704999 | 88706881 | ENSG00000124391 | NA | NA | NA | NA | NA |
| IL18 | IL.18 | Q14116 | Interleukin-18 (IL-18) | IL-18 | 11 | 112013974 | 112034840 | ENSG00000150782 | NA | NA | NA | NA | NA |
| IL18R1 | IL.18R1 | Q13478 | Interleukin-18 receptor 1 (IL-18R1) | IL-18R1 | 2 | 102927989 | 103015218 | ENSG00000115604 | NA | NA | NA | NA | NA |
| IL1A | IL.1.alpha | P01583 | Interleukin-1 alpha (IL-1 alpha) | IL-1 alpha | 2 | 113531492 | 113542167 | ENSG00000115008 | NA | NA | NA | NA | NA |
| IL2 | IL.2 | P60568 | Interleukin-2 (IL-2) | IL-2 | 4 | 123372625 | 123377880 | ENSG00000109471 | NA | NA | NA | NA | NA |
| IL20 | IL.20 | Q9NYY1 | Interleukin-20 (IL-20) | IL-20 | 1 | 207038699 | 207042568 | ENSG00000162891 | NA | NA | NA | NA | NA |
| IL20RA | IL.20RA | Q9UHF4 | Interleukin-20 receptor subunit alpha (IL-20RA) | IL-20RA | 6 | 137321108 | 137366298 | ENSG00000016402 | NA | NA | NA | NA | NA |
| IL22RA1 | IL.22.RA1 | Q8N6P7 | Interleukin-22 receptor subunit alpha-1 (IL-22RA1) | IL-22RA1 | 1 | 24446261 | 24469611 | ENSG00000142677 | NA | NA | NA | NA | NA |
| IL24 | IL.24 | Q13007 | Interleukin-24 (IL-24) | IL-24 | 1 | 207070788 | 207077484 | ENSG00000162892 | NA | NA | NA | NA | NA |
| IL2RB | IL.2RB | P14784 | Interleukin-2 receptor subunit beta (IL-2RB) | IL-2RB | 22 | 37521878 | 37571094 | ENSG00000100385 | NA | NA | NA | NA | NA |
| IL33 | IL.33 | O95760 | Interleukin-33 (IL-33) | IL-33 | 9 | 6215805 | 6257983 | ENSG00000137033 | NA | NA | NA | NA | NA |
| IL4 | IL.4 | P05112 | Interleukin-4 (IL-4) | IL-4 | 5 | 132009678 | 132018368 | ENSG00000113520 | NA | NA | NA | NA | NA |
| IL5 | IL.5 | P05113 | Interleukin-5 (IL-5) | IL-5 | 5 | 131877136 | 131892530 | ENSG00000113525 | NA | NA | NA | NA | NA |
| IL6 | IL.6 | P05231 | Interleukin-6 (IL-6) | IL-6 | 7 | 22765503 | 22771621 | ENSG00000136244 | NA | NA | NA | NA | NA |
| IL7 | IL.7 | P13232 | Interleukin-7 (IL-7) | IL-7 | 8 | 79587978 | 79717758 | ENSG00000104432 | NA | NA | NA | NA | NA |
| IL8 | IL.8 | P10145 | Interleukin-8 (IL-8) | IL-8 | 4 | 74606223 | 74609433 | ENSG00000169429 | NA | NA | NA | NA | NA |
| KITLG | SCF | P21583 | Stem cell factor (SCF) | SCF | 12 | 88886570 | 88974628 | ENSG00000049130 | NA | NA | NA | NA | NA |
| LIF | LIF | P15018 | Leukemia inhibitory factor (LIF) | LIF | 22 | 30636436 | 30642840 | ENSG00000128342 | NA | NA | NA | NA | NA |
| LIFR | LIF.R | P42702 | Leukemia inhibitory factor receptor (LIF-R) | LIF-R | 5 | 38475065 | 38608456 | ENSG00000113594 | NA | NA | NA | NA | NA |
| LTA | TNFB | P01374 | TNF-beta (TNFB) | TNFB | 6 | 31539831 | 31542101 | ENSG00000226979 | NA | NA | NA | NA | NA |
| MMP1 | MMP.1 | P03956 | Matrix metalloproteinase-1 (MMP-1) | MMP-1 | 11 | 102660651 | 102668891 | ENSG00000196611 | NA | NA | NA | NA | NA |
| MMP10 | MMP.10 | P09238 | Matrix metalloproteinase-10 (MMP-10) | MMP-10 | 11 | 102641234 | 102651359 | ENSG00000166670 | NA | NA | NA | NA | NA |
| NGF | Beta.NGF | P01138 | Beta-nerve growth factor (Beta-NGF) | Beta-NGF | 1 | 115828539 | 115880857 | ENSG00000134259 | NA | NA | NA | NA | NA |
| NRTN | NRTN | Q99748 | Neurturin (NRTN) | NRTN | 19 | 5823813 | 5828335 | ENSG00000171119 | NA | NA | NA | NA | NA |
| NTF3 | NT.3 | P20783 | Neurotrophin-3 (NT-3) | NT-3 | 12 | 5541278 | 5630702 | ENSG00000185652 | NA | NA | NA | NA | NA |
| OSM | OSM | P13725 | Oncostatin-M (OSM) | OSM | 22 | 30658818 | 30662829 | ENSG00000099985 | NA | NA | NA | NA | NA |
| PLAU | uPA | P00749 | Urokinase-type plasminogen activator (uPA) | uPA | 10 | 75668935 | 75677255 | ENSG00000122861 | NA | NA | NA | NA | NA |
| S100A12 | EN.RAGE | P80511 | Protein S100-A12 (EN-RAGE) | EN-RAGE | 1 | 153346184 | 153348125 | ENSG00000163221 | NA | NA | NA | NA | NA |
| SIRT2 | SIRT2 | Q8IXJ6 | SIR2-like protein 2 (SIRT2) | SIRT2 | 19 | 39369197 | 39390502 | ENSG00000068903 | NA | NA | NA | NA | NA |
| SLAMF1 | SLAMF1 | Q13291 | Signaling lymphocytic activation molecule (SLAMF1) | SLAMF1 | 1 | 160577890 | 160617085 | ENSG00000117090 | NA | NA | NA | NA | NA |
| STAMBP | STAMPB | O95630 | STAM-binding protein (STAMPB) | STAMPB | 2 | 74056086 | 74100786 | ENSG00000124356 | NA | NA | NA | NA | NA |
| SULT1A1 | ST1A1 | P50225 | Sulfotransferase 1A1 (ST1A1) | ST1A1 | 16 | 28616903 | 28634946 | ENSG00000196502 | NA | NA | NA | NA | NA |
| TGFA | TGF.alpha | P01135 | Transforming growth factor alpha (TGF-alpha) | TGF-alpha | 2 | 70674412 | 70781325 | ENSG00000163235 | NA | NA | NA | NA | NA |
| TGFB1 | LAP.TGF.beta.1 | P01137 | Latency-associated peptide transforming growth factor beta 1 (LAP TGF-beta-1) | LAP TGF-beta-1 | 19 | 41807492 | 41859816 | ENSG00000105329 | NA | NA | NA | NA | NA |
| TNF | TNF | P01375 | Tumor necrosis factor (TNF) | TNF | 6 | 31543344 | 31546113 | ENSG00000232810 | NA | NA | NA | NA | NA |
| TNFRSF11B | OPG | O00300 | Osteoprotegerin (OPG) | OPG | 8 | 119935796 | 119964439 | ENSG00000164761 | NA | NA | NA | NA | NA |
| TNFRSF9 | TNFRSF9 | Q07011 | Tumor necrosis factor receptor superfamily member 9 (TNFRSF9) | TNFRSF9 | 1 | 7979907 | 8000926 | ENSG00000049249 | NA | NA | NA | NA | NA |
| TNFSF10 | TRAIL | P50591 | TNF-related apoptosis ligand (TRAIL) | TRAIL | 3 | 172223298 | 172241297 | ENSG00000121858 | NA | NA | NA | NA | NA |
| TNFSF11 | TRANCE | O14788 | TNF-related activation cytokine (TRANCE) | TRANCE | 13 | 43136872 | 43182149 | ENSG00000120659 | NA | NA | NA | NA | NA |
| TNFSF12 | TWEAK | O43508 | Tumor necrosis factor (Ligand) superfamily member 12 (TWEAK) | TWEAK | 17 | 7452208 | 7464925 | ENSG00000239697 | NA | NA | NA | NA | NA |
| TNFSF14 | TNFSF14 | O43557 | Tumor necrosis factor ligand superfamily member 14 (TNFSF14) | TNFSF14 | 19 | 6663148 | 6670599 | ENSG00000125735 | NA | ENSG00000125735 | 19 | 6661253 | 6670588 |
| TSLP | TSLP | Q969D9 | Thymic stromal lymphopoietin (TSLP) | TSLP | 5 | 110405760 | 110413722 | ENSG00000145777 | NA | NA | NA | NA | NA |
| VEGFA | VEGF.A | P15692 | Vascular endothelial growth factor A (VEGF_A) | VEGF_A | 6 | 43737921 | 43754224 | ENSG00000112715 | NA | NA | NA | NA | NA |
4 Reference data
We showcase EnsDb.Hsapiens.v75 from Bioconductor.
options(new_options)
ensembldb::metadata(EnsDb.Hsapiens.v75)
#> name value
#> 1 Db type EnsDb
#> 2 Type of Gene ID Ensembl Gene ID
#> 3 Supporting package ensembldb
#> 4 Db created by ensembldb package from Bioconductor
#> 5 script_version 0.3.0
#> 6 Creation time Thu May 18 09:15:45 2017
#> 7 ensembl_version 75
#> 8 ensembl_host localhost
#> 9 Organism homo_sapiens
#> 10 taxonomy_id 9606
#> 11 genome_build GRCh37
#> 12 DBSCHEMAVERSION 2.0
ensembldb::keytypes(EnsDb.Hsapiens.v75)
#> [1] "ENTREZID" "EXONID" "GENEBIOTYPE"
#> [4] "GENEID" "GENENAME" "PROTDOMID"
#> [7] "PROTEINDOMAINID" "PROTEINDOMAINSOURCE" "PROTEINID"
#> [10] "SEQNAME" "SEQSTRAND" "SYMBOL"
#> [13] "TXBIOTYPE" "TXID" "TXNAME"
#> [16] "UNIPROTID"
exon_info <- ensembldb::exons(EnsDb.Hsapiens.v75)
gene_info <- ensembldb::genes(EnsDb.Hsapiens.v75)
transcript_info <- ensembldb::transcripts(EnsDb.Hsapiens.v75)
colnames(S4Vectors::mcols(gene_info))
#> [1] "gene_id" "gene_name" "gene_biotype" "seq_coord_system"
#> [5] "symbol" "entrezid"
colnames(S4Vectors::mcols(transcript_info))
#> [1] "tx_id" "tx_biotype" "tx_cds_seq_start" "tx_cds_seq_end"
#> [5] "gene_id" "tx_name"
overlaps <- IRanges::findOverlaps(transcript_info, gene_info)
overlapping_transcripts <- transcript_info[queryHits(overlaps)]
overlapping_genes <- gene_info[subjectHits(overlaps)]
overlap_data <- data.frame(
transcript_id = mcols(overlapping_transcripts)$tx_id,
gene_id = S4Vectors::mcols(overlapping_genes)$gene_id,
gene_name = S4Vectors::mcols(overlapping_genes)$gene_name,
start = pmax(start(overlapping_transcripts), start(overlapping_genes)),
end = pmin(end(overlapping_transcripts), end(overlapping_genes))
)
gene_symbols <- c("BRCA1", "TP53")
gene_data <- subset(overlap_data,gene_name%in%gene_symbols,select=-c(gene_id,gene_name))
cols <- c("UNIPROTID","PROTEINID","GENEID","GENENAME","SEQNAME","TXID")
info <- ensembldb::select(EnsDb.Hsapiens.v75, keys = gene_symbols,
columns = cols, keytype = "SYMBOL") |>
dplyr::left_join(head(gene_data,15),by=c('TXID'='transcript_id')) |>
subset(!is.na(UNIPROTID)&!is.na(start)&!is.na(end))
knitr::kable(head(info,15),caption="Annotation for BRCA1 and TP53")| SYMBOL | UNIPROTID | PROTEINID | GENEID | GENENAME | SEQNAME | TXID | start | end | |
|---|---|---|---|---|---|---|---|---|---|
| 242 | TP53 | Q761V2_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 243 | TP53 | Q6IT77_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 244 | TP53 | Q1HGV1_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 245 | TP53 | Q0PKT5_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 246 | TP53 | L0ES54_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 247 | TP53 | G4Y083_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 248 | TP53 | E9PCY9_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 249 | TP53 | E7EQX7_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 250 | TP53 | E7EMR6_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 251 | TP53 | B5AKF6_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 252 | TP53 | A4GWD0_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 253 | TP53 | A4GWB8_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 254 | TP53 | A4GWB5_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 255 | TP53 | A2I9Z0_HUMAN | ENSP00000410739 | ENSG00000141510 | TP53 | 17 | ENST00000413465 | 7565097 | 7579912 |
| 256 | TP53 | S5LQU8_HUMAN | ENSP00000352610 | ENSG00000141510 | TP53 | 17 | ENST00000359597 | 7569404 | 7579912 |
keytypes(org.Hs.eg.db)
#> [1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
#> [6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
#> [11] "GENETYPE" "GO" "GOALL" "IPI" "MAP"
#> [16] "OMIM" "ONTOLOGY" "ONTOLOGYALL" "PATH" "PFAM"
#> [21] "PMID" "PROSITE" "REFSEQ" "SYMBOL" "UCSCKG"
#> [26] "UNIPROT"
uniprot_ids <- ensembldb::select(org.Hs.eg.db, keys = gene_symbols, columns = "UNIPROT", keytype = "SYMBOL")
#> 'select()' returned 1:many mapping between keys and columns
options(sys_options)where org.Hs.eg.db is more focused on genes.
5 Scripts
An analysis involving COVID-19 data is in Olink/ directory, while the scripts/ directory records data generation which potentially can be extended.
Specifically, docs.sh operates with GitHub while cran.sh builds, installs, and checks for compliance with the Comprehensive R Archive Network (CRAN).
6 URLs
- CellCarta, https://cellcarta.com/
- EndNote, https://endnote.com/
- ENSEMBL BioMart, https://www.ensembl.org/index.html
- GitHub, https://github.com/
- NULISA, https://alamarbio.com/technology/nulisa-platform/
- NCI Proteomic Data Commons, https://pdc.cancer.gov/pdc/browse
- Olink, https://olink.com/ (https://olink.com/publications)
- SCALLOP consortium, http://www.scallop-consortium.com/
- Seer, https://seer.bio/
- SomaLogic, https://somalogic.com/
- SWATH-MS, https://www.creative-proteomics.com/ngpro/swath-ms.html
- Thermo Fisher Scientific - LSMS, https://github.com/thermofisherlsms
- UCSC, https://genome.ucsc.edu/
References
The EndNote/ directory includes references in1 and4 formatted in EndNote.