It aggregates protein panel data and metadata for protein quantitative trait locus (pQTL) analysis using 'pQTLtools' (https://jinghuazhao.github.io/pQTLtools/). The package includes data from affinity-based panels such as 'Olink' (https://olink.com/) and 'SomaScan' (https://somalogic.com/), as well as mass spectrometry-based panels from 'CellCarta' (https://cellcarta.com/), 'Seer' (https://seer.bio/) and 'SWATH-MS' (doi:10.15252/msb.20178126 ). The metadata encompasses updated annotations and publication details.
Details
Available data are listed in the following table.
| Objects | Description |
| Datasets | |
caprion | Caprion panel |
inf1 | Olink/INF panel |
Olink_Explore_1536 | Olink/NGS 1472 panels |
Olink_Explore_3072 | Olink/Explore 3072 panels |
Olink_Explore_HT | Olink/Explore HT panels |
Olink_Target_96 | Olink/Target 96 panels |
Olink_qPCR | Olink/qPCR panels |
SomaScan160410 | SomaScan panel |
SomaScanV4.1 | SomaScan v4.1 panel |
SomaScan11k | SomaScan 11k panel |
scallop_inf1 | SCALLOP/INF meta-analysis results |
seer1980 | ST1 from Suhre et al. (2024) |
swath_ms | A curated SWATH-MS panel, see Ludwig et al. (2018) |
| Installations | |
| EndNote/ | Proteogenomics references |
| Olink/ | Olink-COVID analysis by MGH |
Some generic description for the datasets are as follows.
chr Chromosome.
start Start position.
end End position.
gene Gene name.
UniProt UniProt ID.
References
Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC, Aebersold R (2018).
“Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial.”
Molecular Systems Biology, 14(8), e8126.
doi:10.15252/msb.20178126
.
Suhre K, Chen Q, Halama A, Mendez K, Dahlin A, Stephan N, Thareja G, Sarwath H, Guturu H, Dwaraka VB, Batzoglou S, Schmidt F, Lasky-Su JA (2024).
“A genome-wide association study of mass spectrometry proteomics using the Seer Proteograph platform.”
BioRxiv.
doi:10.1101/2024.05.27.596028
.
Examples
# \donttest{
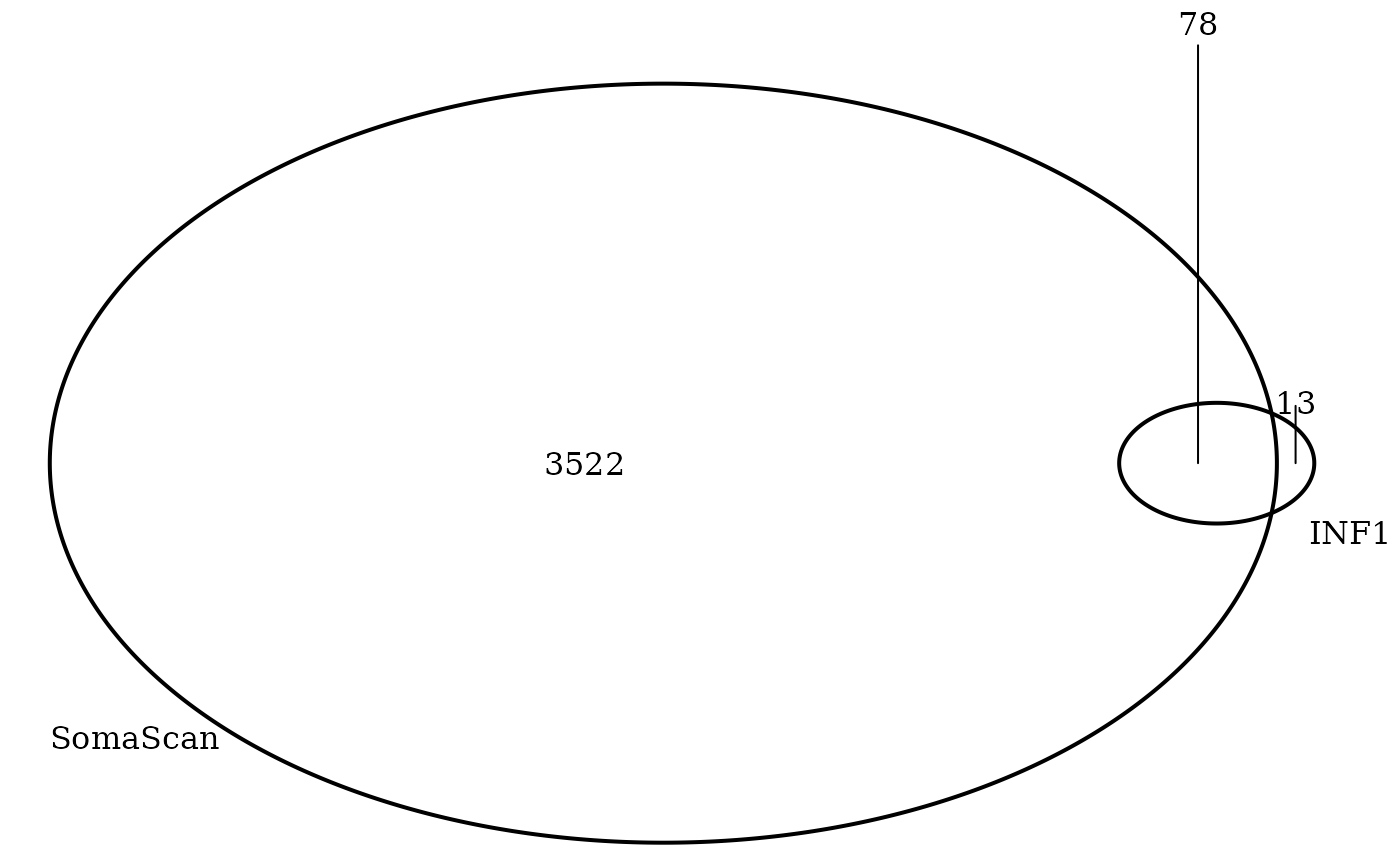
# Olink-SomaScan panel overlap
p <- list(setdiff(inf1$uniprot,"P23560"),
setdiff(SomaScan160410$UniProt[!is.na(SomaScan160410$UniProt)],"P23560"))
cnames <- c("INF1","SomaScan")
os <- VennDiagram::venn.diagram(x = p, category.names=cnames, filename=NULL,
disable.logging = TRUE,height=8,width=8,units="in")
#> INFO [2025-03-26 17:44:50] $x
#> INFO [2025-03-26 17:44:50] p
#> INFO [2025-03-26 17:44:50]
#> INFO [2025-03-26 17:44:50] $category.names
#> INFO [2025-03-26 17:44:50] cnames
#> INFO [2025-03-26 17:44:50]
#> INFO [2025-03-26 17:44:50] $filename
#> INFO [2025-03-26 17:44:50] NULL
#> INFO [2025-03-26 17:44:50]
#> INFO [2025-03-26 17:44:50] $disable.logging
#> INFO [2025-03-26 17:44:50] [1] TRUE
#> INFO [2025-03-26 17:44:50]
#> INFO [2025-03-26 17:44:50] $height
#> INFO [2025-03-26 17:44:50] [1] 8
#> INFO [2025-03-26 17:44:50]
#> INFO [2025-03-26 17:44:50] $width
#> INFO [2025-03-26 17:44:50] [1] 8
#> INFO [2025-03-26 17:44:50]
#> INFO [2025-03-26 17:44:50] $units
#> INFO [2025-03-26 17:44:50] [1] "in"
#> INFO [2025-03-26 17:44:50]
grid::grid.newpage()
grid::grid.draw(os)
 m <- merge(inf1,SomaScan160410,by.x="uniprot",by.y="UniProt")
u <- setdiff(with(m,unique(uniprot)),"P23560")
o <- subset(inf1,uniprot %in% u)
dim(o)
#> [1] 78 14
vars <- c("UniProt","chr","start","end","extGene","Target","TargetFullName")
s <- subset(SomaScan160410[vars], UniProt %in% u)
dim(s)
#> [1] 127 7
us <- s[!duplicated(s),]
dim(us)
#> [1] 95 7
us
#> UniProt chr start end extGene Target
#> 110 Q13651 11 117857063 117872196 IL10RA IL-10 Ra
#> 127 P29460 5 158741791 158757895 IL12B IL-23
#> 131 P29460 5 158741791 158757895 IL12B IL-12
#> 361 P05113 5 131877136 131892530 IL5 IL-5
#> 541 Q8NFT8 2 230222345 230579274 DNER DNER
#> 605 Q07325 4 76922428 76928641 CXCL9 MIG
#> 1077 O95760 9 6215805 6257983 IL33 IL33
#> 1352 P28325 20 23856572 23860387 CST5 CYTD
#> 1385 Q16552 6 52051185 52055436 IL17A IL-17
#> 1476 P15692 6 43737921 43754224 VEGFA VEGF121
#> 1487 P13232 8 79587978 79717758 IL7 IL-7
#> 1490 Q13261 10 5990855 6020150 IL15RA IL-15 Ra
#> 1494 P13725 22 30658818 30662829 OSM OSM
#> 1496 P06127 11 60869867 60895324 CD5 CD5
#> 1499 O15444 19 8117651 8127534 CCL25 TECK
#> 1505 Q13478 2 102927989 103015218 IL18R1 IL-18 Ra
#> 1515 P49771 19 49977464 49989488 FLT3LG Flt3 ligand
#> 1596 P01579 12 68548548 68553527 IFNG IFN-g
#> 1697 P01137 19 41807492 41859816 TGFB1 TGF-b1
#> 1713 P78556 2 228678558 228682272 CCL20 MIP-3a
#> 1734 O00300 8 119935796 119964439 TNFRSF11B OPG
#> 1739 P05231 7 22765503 22771621 IL6 IL-6
#> 1741 P13500 17 32582304 32584222 CCL2 MCP-1
#> 1771 P15692 6 43737921 43754224 VEGFA VEGF
#> 1772 Q07011 1 7979907 8000926 TNFRSF9 4-1BB
#> 1795 Q08334 21 34638663 34669539 IL10RB IL-10 Rb
#> 1822 P14210 7 81328322 81399754 HGF HGF
#> 1873 O95750 11 69513000 69519410 FGF19 FGF-19
#> 1879 P22301 1 206940947 206945839 IL10 IL-10
#> 1891 P80075 17 32646055 32648421 CCL8 MCP-2
#> 1913 P78423 16 57406370 57418960 CX3CL1 Fractalkine/CX3CL-1
#> 1969 Q9NRJ3 5 43376747 43412493 CCL28 CCL28
#> 1972 P05112 5 132009678 132018368 IL4 IL-4
#> 1974 P55773 17 34340096 34345005 CCL23 MPIF-1
#> 1979 O14788 13 43136872 43182149 TNFSF11 sRANKL
#> 1983 Q5T4W7 1 44398992 44402913 ARTN Artemin
#> 2020 P42830 4 74861359 74864496 CXCL5 ENA-78
#> 2023 P09341 4 74735110 74736959 CXCL1 Gro-a
#> 2049 Q969D9 5 110405760 110413722 TSLP TSLP
#> 2054 P55773 17 34340096 34345005 CCL23 Ck-b-8-1
#> 2073 O14625 4 76954835 76962568 CXCL11 I-TAC
#> 2074 P10147 17 34415602 34417515 CCL3 MIP-1a
#> 2100 P12034 4 81187753 81257834 FGF5 FGF-5
#> 2103 P60568 4 123372625 123377880 IL2 IL-2
#> 2104 P35225 5 131991955 131996802 IL13 IL-13
#> 2219 Q13007 1 207070788 207077484 IL24 IL24
#> 2317 P10145 4 74606223 74609433 IL8 IL-8
#> 2351 P80162 4 74702214 74714781 CXCL6 GCP-2
#> 2372 P01374 6 31539831 31542101 LTA Lymphotoxin a1/b2
#> 2373 P01374 6 31529774 31532044 LTA Lymphotoxin a1/b2
#> 2374 P01374 6 31579318 31581588 LTA Lymphotoxin a1/b2
#> 2383 P01374 6 31539831 31542101 LTA Lymphotoxin a2/b1
#> 2384 P01374 6 31529774 31532044 LTA Lymphotoxin a2/b1
#> 2385 P01374 6 31579318 31581588 LTA Lymphotoxin a2/b1
#> 2431 Q8N6P7 1 24446261 24469611 IL22RA1 IL22RA1
#> 2476 P09603 1 110452864 110473614 CSF1 CSF-1
#> 2480 P09238 11 102641234 102651359 MMP10 MMP-10
#> 2497 Q9GZV9 12 4477393 4488894 FGF23 FGF23
#> 2600 Q9NYY1 1 207038699 207042568 IL20 IL-20
#> 2603 P02778 4 76942273 76944650 CXCL10 IP-10
#> 2605 Q99616 17 32683471 32685629 CCL13 MCP-4
#> 2606 P20783 12 5541278 5630702 NTF3 Neurotrophin-3
#> 2627 P00749 10 75668935 75677255 PLAU uPA
#> 2806 P01374 6 31539831 31542101 LTA TNF-b
#> 2807 P01374 6 31529774 31532044 LTA TNF-b
#> 2808 P01374 6 31579318 31581588 LTA TNF-b
#> 2840 P01583 2 113531492 113542167 IL1A IL-1a
#> 2856 P80098 17 32597240 32599261 CCL7 MCP-3
#> 2896 Q99731 9 34689564 34691274 CCL19 MIP-3b
#> 2902 P03956 11 102660651 102668891 MMP1 MMP-1
#> 2984 Q8IXJ6 19 39369197 39390502 SIRT2 SIRT2
#> 2996 Q9NZQ7 9 5450503 5470566 CD274 B7-H1
#> 2999 Q9BZW8 1 160799950 160832692 CD244 CD244
#> 3017 Q9UHF4 6 137321108 137366298 IL20RA IL-20 Ra
#> 3152 P51671 17 32612687 32615353 CCL11 Eotaxin
#> 3173 O43557 19 6663148 6670599 TNFSF14 LIGHT
#> 3402 P01138 1 115828539 115880857 NGF b-NGF
#> 3407 P39905 5 37812779 37839788 GDNF GDNF
#> 3410 P42702 5 38475065 38608456 LIFR LIF sR
#> 3418 P80511 1 153346184 153348125 S100A12 S100A12
#> 3445 P01375 6 31530756 31533525 TNF TNF-a
#> 3446 P01375 6 31533585 31536356 TNF TNF-a
#> 3447 P01375 6 31619653 31622422 TNF TNF-a
#> 3448 P01375 6 31582831 31585600 TNF TNF-a
#> 3449 P01375 6 31525488 31528257 TNF TNF-a
#> 3450 P01375 6 31533287 31536056 TNF TNF-a
#> 3451 P01375 6 31543344 31546113 TNF TNF-a
#> 3452 O43508 17 7452208 7464925 TNFSF12 TWEAK
#> 3692 Q9H5V8 3 45123770 45187914 CDCP1 CDCP1
#> 4021 P15018 22 30636436 30642840 LIF LIF
#> 4159 Q13291 1 160577890 160617085 SLAMF1 SLAF1
#> 4766 Q9P0M4 16 88704999 88706881 IL17C IL-17C
#> 4828 P14784 22 37521878 37571094 IL2RB IL-2 sRb
#> 4836 P01135 2 70674412 70781325 TGFA TGF-a
#> 4850 P21583 12 88886570 88974628 KITLG SCF
#> TargetFullName
#> 110 Interleukin-10 receptor subunit alpha
#> 127 Interleukin-23
#> 131 Interleukin-12
#> 361 Interleukin-5
#> 541 Delta and Notch-like epidermal growth factor-related receptor
#> 605 C-X-C motif chemokine 9
#> 1077 Interleukin-33
#> 1352 Cystatin-D
#> 1385 Interleukin-17A
#> 1476 Vascular endothelial growth factor A, isoform 121
#> 1487 Interleukin-7
#> 1490 Interleukin-15 receptor subunit alpha
#> 1494 Oncostatin-M
#> 1496 T-cell surface glycoprotein CD5
#> 1499 C-C motif chemokine 25
#> 1505 Interleukin-18 receptor 1
#> 1515 Fms-related tyrosine kinase 3 ligand
#> 1596 Interferon gamma
#> 1697 Transforming growth factor beta-1
#> 1713 C-C motif chemokine 20
#> 1734 Tumor necrosis factor receptor superfamily member 11B
#> 1739 Interleukin-6
#> 1741 C-C motif chemokine 2
#> 1771 Vascular endothelial growth factor A
#> 1772 Tumor necrosis factor receptor superfamily member 9
#> 1795 Interleukin-10 receptor subunit beta
#> 1822 Hepatocyte growth factor
#> 1873 Fibroblast growth factor 19
#> 1879 Interleukin-10
#> 1891 C-C motif chemokine 8
#> 1913 Fractalkine
#> 1969 C-C motif chemokine 28
#> 1972 Interleukin-4
#> 1974 C-C motif chemokine 23
#> 1979 Tumor necrosis factor ligand superfamily member 11
#> 1983 Artemin
#> 2020 C-X-C motif chemokine 5
#> 2023 Growth-regulated alpha protein
#> 2049 Thymic stromal lymphopoietin
#> 2054 Ck-beta-8-1
#> 2073 C-X-C motif chemokine 11
#> 2074 C-C motif chemokine 3
#> 2100 Fibroblast growth factor 5
#> 2103 Interleukin-2
#> 2104 Interleukin-13
#> 2219 Interleukin-24
#> 2317 Interleukin-8
#> 2351 C-X-C motif chemokine 6
#> 2372 Lymphotoxin alpha1:beta2
#> 2373 Lymphotoxin alpha1:beta2
#> 2374 Lymphotoxin alpha1:beta2
#> 2383 Lymphotoxin alpha2:beta1
#> 2384 Lymphotoxin alpha2:beta1
#> 2385 Lymphotoxin alpha2:beta1
#> 2431 Interleukin-22 receptor subunit alpha-1
#> 2476 Macrophage colony-stimulating factor 1
#> 2480 Stromelysin-2
#> 2497 Fibroblast growth factor 23
#> 2600 Interleukin-20
#> 2603 C-X-C motif chemokine 10
#> 2605 C-C motif chemokine 13
#> 2606 Neurotrophin-3
#> 2627 Urokinase-type plasminogen activator
#> 2806 Lymphotoxin-alpha
#> 2807 Lymphotoxin-alpha
#> 2808 Lymphotoxin-alpha
#> 2840 Interleukin-1 alpha
#> 2856 C-C motif chemokine 7
#> 2896 C-C motif chemokine 19
#> 2902 Interstitial collagenase
#> 2984 NAD-dependent protein deacetylase sirtuin-2
#> 2996 Programmed cell death 1 ligand 1
#> 2999 Natural killer cell receptor 2B4
#> 3017 Interleukin-20 receptor subunit alpha
#> 3152 Eotaxin
#> 3173 Tumor necrosis factor ligand superfamily member 14
#> 3402 beta-nerve growth factor
#> 3407 Glial cell line-derived neurotrophic factor
#> 3410 Leukemia inhibitory factor receptor
#> 3418 Protein S100-A12
#> 3445 Tumor necrosis factor
#> 3446 Tumor necrosis factor
#> 3447 Tumor necrosis factor
#> 3448 Tumor necrosis factor
#> 3449 Tumor necrosis factor
#> 3450 Tumor necrosis factor
#> 3451 Tumor necrosis factor
#> 3452 Tumor necrosis factor ligand superfamily member 12
#> 3692 CUB domain-containing protein 1
#> 4021 Leukemia inhibitory factor
#> 4159 Signaling lymphocytic activation molecule
#> 4766 Interleukin-17C
#> 4828 Interleukin-2 receptor subunit beta
#> 4836 Transforming growth factor alpha
#> 4850 Kit ligand
# }
m <- merge(inf1,SomaScan160410,by.x="uniprot",by.y="UniProt")
u <- setdiff(with(m,unique(uniprot)),"P23560")
o <- subset(inf1,uniprot %in% u)
dim(o)
#> [1] 78 14
vars <- c("UniProt","chr","start","end","extGene","Target","TargetFullName")
s <- subset(SomaScan160410[vars], UniProt %in% u)
dim(s)
#> [1] 127 7
us <- s[!duplicated(s),]
dim(us)
#> [1] 95 7
us
#> UniProt chr start end extGene Target
#> 110 Q13651 11 117857063 117872196 IL10RA IL-10 Ra
#> 127 P29460 5 158741791 158757895 IL12B IL-23
#> 131 P29460 5 158741791 158757895 IL12B IL-12
#> 361 P05113 5 131877136 131892530 IL5 IL-5
#> 541 Q8NFT8 2 230222345 230579274 DNER DNER
#> 605 Q07325 4 76922428 76928641 CXCL9 MIG
#> 1077 O95760 9 6215805 6257983 IL33 IL33
#> 1352 P28325 20 23856572 23860387 CST5 CYTD
#> 1385 Q16552 6 52051185 52055436 IL17A IL-17
#> 1476 P15692 6 43737921 43754224 VEGFA VEGF121
#> 1487 P13232 8 79587978 79717758 IL7 IL-7
#> 1490 Q13261 10 5990855 6020150 IL15RA IL-15 Ra
#> 1494 P13725 22 30658818 30662829 OSM OSM
#> 1496 P06127 11 60869867 60895324 CD5 CD5
#> 1499 O15444 19 8117651 8127534 CCL25 TECK
#> 1505 Q13478 2 102927989 103015218 IL18R1 IL-18 Ra
#> 1515 P49771 19 49977464 49989488 FLT3LG Flt3 ligand
#> 1596 P01579 12 68548548 68553527 IFNG IFN-g
#> 1697 P01137 19 41807492 41859816 TGFB1 TGF-b1
#> 1713 P78556 2 228678558 228682272 CCL20 MIP-3a
#> 1734 O00300 8 119935796 119964439 TNFRSF11B OPG
#> 1739 P05231 7 22765503 22771621 IL6 IL-6
#> 1741 P13500 17 32582304 32584222 CCL2 MCP-1
#> 1771 P15692 6 43737921 43754224 VEGFA VEGF
#> 1772 Q07011 1 7979907 8000926 TNFRSF9 4-1BB
#> 1795 Q08334 21 34638663 34669539 IL10RB IL-10 Rb
#> 1822 P14210 7 81328322 81399754 HGF HGF
#> 1873 O95750 11 69513000 69519410 FGF19 FGF-19
#> 1879 P22301 1 206940947 206945839 IL10 IL-10
#> 1891 P80075 17 32646055 32648421 CCL8 MCP-2
#> 1913 P78423 16 57406370 57418960 CX3CL1 Fractalkine/CX3CL-1
#> 1969 Q9NRJ3 5 43376747 43412493 CCL28 CCL28
#> 1972 P05112 5 132009678 132018368 IL4 IL-4
#> 1974 P55773 17 34340096 34345005 CCL23 MPIF-1
#> 1979 O14788 13 43136872 43182149 TNFSF11 sRANKL
#> 1983 Q5T4W7 1 44398992 44402913 ARTN Artemin
#> 2020 P42830 4 74861359 74864496 CXCL5 ENA-78
#> 2023 P09341 4 74735110 74736959 CXCL1 Gro-a
#> 2049 Q969D9 5 110405760 110413722 TSLP TSLP
#> 2054 P55773 17 34340096 34345005 CCL23 Ck-b-8-1
#> 2073 O14625 4 76954835 76962568 CXCL11 I-TAC
#> 2074 P10147 17 34415602 34417515 CCL3 MIP-1a
#> 2100 P12034 4 81187753 81257834 FGF5 FGF-5
#> 2103 P60568 4 123372625 123377880 IL2 IL-2
#> 2104 P35225 5 131991955 131996802 IL13 IL-13
#> 2219 Q13007 1 207070788 207077484 IL24 IL24
#> 2317 P10145 4 74606223 74609433 IL8 IL-8
#> 2351 P80162 4 74702214 74714781 CXCL6 GCP-2
#> 2372 P01374 6 31539831 31542101 LTA Lymphotoxin a1/b2
#> 2373 P01374 6 31529774 31532044 LTA Lymphotoxin a1/b2
#> 2374 P01374 6 31579318 31581588 LTA Lymphotoxin a1/b2
#> 2383 P01374 6 31539831 31542101 LTA Lymphotoxin a2/b1
#> 2384 P01374 6 31529774 31532044 LTA Lymphotoxin a2/b1
#> 2385 P01374 6 31579318 31581588 LTA Lymphotoxin a2/b1
#> 2431 Q8N6P7 1 24446261 24469611 IL22RA1 IL22RA1
#> 2476 P09603 1 110452864 110473614 CSF1 CSF-1
#> 2480 P09238 11 102641234 102651359 MMP10 MMP-10
#> 2497 Q9GZV9 12 4477393 4488894 FGF23 FGF23
#> 2600 Q9NYY1 1 207038699 207042568 IL20 IL-20
#> 2603 P02778 4 76942273 76944650 CXCL10 IP-10
#> 2605 Q99616 17 32683471 32685629 CCL13 MCP-4
#> 2606 P20783 12 5541278 5630702 NTF3 Neurotrophin-3
#> 2627 P00749 10 75668935 75677255 PLAU uPA
#> 2806 P01374 6 31539831 31542101 LTA TNF-b
#> 2807 P01374 6 31529774 31532044 LTA TNF-b
#> 2808 P01374 6 31579318 31581588 LTA TNF-b
#> 2840 P01583 2 113531492 113542167 IL1A IL-1a
#> 2856 P80098 17 32597240 32599261 CCL7 MCP-3
#> 2896 Q99731 9 34689564 34691274 CCL19 MIP-3b
#> 2902 P03956 11 102660651 102668891 MMP1 MMP-1
#> 2984 Q8IXJ6 19 39369197 39390502 SIRT2 SIRT2
#> 2996 Q9NZQ7 9 5450503 5470566 CD274 B7-H1
#> 2999 Q9BZW8 1 160799950 160832692 CD244 CD244
#> 3017 Q9UHF4 6 137321108 137366298 IL20RA IL-20 Ra
#> 3152 P51671 17 32612687 32615353 CCL11 Eotaxin
#> 3173 O43557 19 6663148 6670599 TNFSF14 LIGHT
#> 3402 P01138 1 115828539 115880857 NGF b-NGF
#> 3407 P39905 5 37812779 37839788 GDNF GDNF
#> 3410 P42702 5 38475065 38608456 LIFR LIF sR
#> 3418 P80511 1 153346184 153348125 S100A12 S100A12
#> 3445 P01375 6 31530756 31533525 TNF TNF-a
#> 3446 P01375 6 31533585 31536356 TNF TNF-a
#> 3447 P01375 6 31619653 31622422 TNF TNF-a
#> 3448 P01375 6 31582831 31585600 TNF TNF-a
#> 3449 P01375 6 31525488 31528257 TNF TNF-a
#> 3450 P01375 6 31533287 31536056 TNF TNF-a
#> 3451 P01375 6 31543344 31546113 TNF TNF-a
#> 3452 O43508 17 7452208 7464925 TNFSF12 TWEAK
#> 3692 Q9H5V8 3 45123770 45187914 CDCP1 CDCP1
#> 4021 P15018 22 30636436 30642840 LIF LIF
#> 4159 Q13291 1 160577890 160617085 SLAMF1 SLAF1
#> 4766 Q9P0M4 16 88704999 88706881 IL17C IL-17C
#> 4828 P14784 22 37521878 37571094 IL2RB IL-2 sRb
#> 4836 P01135 2 70674412 70781325 TGFA TGF-a
#> 4850 P21583 12 88886570 88974628 KITLG SCF
#> TargetFullName
#> 110 Interleukin-10 receptor subunit alpha
#> 127 Interleukin-23
#> 131 Interleukin-12
#> 361 Interleukin-5
#> 541 Delta and Notch-like epidermal growth factor-related receptor
#> 605 C-X-C motif chemokine 9
#> 1077 Interleukin-33
#> 1352 Cystatin-D
#> 1385 Interleukin-17A
#> 1476 Vascular endothelial growth factor A, isoform 121
#> 1487 Interleukin-7
#> 1490 Interleukin-15 receptor subunit alpha
#> 1494 Oncostatin-M
#> 1496 T-cell surface glycoprotein CD5
#> 1499 C-C motif chemokine 25
#> 1505 Interleukin-18 receptor 1
#> 1515 Fms-related tyrosine kinase 3 ligand
#> 1596 Interferon gamma
#> 1697 Transforming growth factor beta-1
#> 1713 C-C motif chemokine 20
#> 1734 Tumor necrosis factor receptor superfamily member 11B
#> 1739 Interleukin-6
#> 1741 C-C motif chemokine 2
#> 1771 Vascular endothelial growth factor A
#> 1772 Tumor necrosis factor receptor superfamily member 9
#> 1795 Interleukin-10 receptor subunit beta
#> 1822 Hepatocyte growth factor
#> 1873 Fibroblast growth factor 19
#> 1879 Interleukin-10
#> 1891 C-C motif chemokine 8
#> 1913 Fractalkine
#> 1969 C-C motif chemokine 28
#> 1972 Interleukin-4
#> 1974 C-C motif chemokine 23
#> 1979 Tumor necrosis factor ligand superfamily member 11
#> 1983 Artemin
#> 2020 C-X-C motif chemokine 5
#> 2023 Growth-regulated alpha protein
#> 2049 Thymic stromal lymphopoietin
#> 2054 Ck-beta-8-1
#> 2073 C-X-C motif chemokine 11
#> 2074 C-C motif chemokine 3
#> 2100 Fibroblast growth factor 5
#> 2103 Interleukin-2
#> 2104 Interleukin-13
#> 2219 Interleukin-24
#> 2317 Interleukin-8
#> 2351 C-X-C motif chemokine 6
#> 2372 Lymphotoxin alpha1:beta2
#> 2373 Lymphotoxin alpha1:beta2
#> 2374 Lymphotoxin alpha1:beta2
#> 2383 Lymphotoxin alpha2:beta1
#> 2384 Lymphotoxin alpha2:beta1
#> 2385 Lymphotoxin alpha2:beta1
#> 2431 Interleukin-22 receptor subunit alpha-1
#> 2476 Macrophage colony-stimulating factor 1
#> 2480 Stromelysin-2
#> 2497 Fibroblast growth factor 23
#> 2600 Interleukin-20
#> 2603 C-X-C motif chemokine 10
#> 2605 C-C motif chemokine 13
#> 2606 Neurotrophin-3
#> 2627 Urokinase-type plasminogen activator
#> 2806 Lymphotoxin-alpha
#> 2807 Lymphotoxin-alpha
#> 2808 Lymphotoxin-alpha
#> 2840 Interleukin-1 alpha
#> 2856 C-C motif chemokine 7
#> 2896 C-C motif chemokine 19
#> 2902 Interstitial collagenase
#> 2984 NAD-dependent protein deacetylase sirtuin-2
#> 2996 Programmed cell death 1 ligand 1
#> 2999 Natural killer cell receptor 2B4
#> 3017 Interleukin-20 receptor subunit alpha
#> 3152 Eotaxin
#> 3173 Tumor necrosis factor ligand superfamily member 14
#> 3402 beta-nerve growth factor
#> 3407 Glial cell line-derived neurotrophic factor
#> 3410 Leukemia inhibitory factor receptor
#> 3418 Protein S100-A12
#> 3445 Tumor necrosis factor
#> 3446 Tumor necrosis factor
#> 3447 Tumor necrosis factor
#> 3448 Tumor necrosis factor
#> 3449 Tumor necrosis factor
#> 3450 Tumor necrosis factor
#> 3451 Tumor necrosis factor
#> 3452 Tumor necrosis factor ligand superfamily member 12
#> 3692 CUB domain-containing protein 1
#> 4021 Leukemia inhibitory factor
#> 4159 Signaling lymphocytic activation molecule
#> 4766 Interleukin-17C
#> 4828 Interleukin-2 receptor subunit beta
#> 4836 Transforming growth factor alpha
#> 4850 Kit ligand
# }