This article collects notes on Bioconductor packages, made available here to faciliate their use and extensions.
pkgs <- c("AnnotationDbi", "AnnotationFilter", "ComplexHeatmap", "DESeq2", "EnsDb.Hsapiens.v86",
"FlowSorted.DLPFC.450k", "GeneNet", "GenomicFeatures", "IlluminaHumanMethylation450kmanifest",
"OUTRIDER","RColorBrewer", "RMariaDB", "Rgraphviz", "S4Vectors", "SummarizedExperiment",
"TxDb.Hsapiens.UCSC.hg38.knownGene", "bladderbatch", "clusterProfiler",
"corpcor", "doParallel", "ensembldb", "fdrtool", "graph", "graphite", "heatmaply",
"minfi", "org.Hs.eg.db", "plyr", "quantro", "recount3", "sva")
es_pkgs <- c("Biobase", "arrayQualityMetrics", "dplyr", "knitr", "mclust", "pQTLtools", "rgl", "scatterplot3d")
se_pkgs <- c("BiocGenerics", "GenomicRanges", "IRanges", "MsCoreUtils", "SummarizedExperiment", "impute")
sp_pkgs <- c("Biostrings", "CAMERA", "MSnbase", "MSstats", "Spectra", "mzR", "protViz", "rawrr")
pkgs <- c(pkgs, es_pkgs,se_pkgs,sp_pkgs)
for (p in pkgs) if (length(grep(paste("^package:", p, "$", sep=""), search())) == 0) {
if (!requireNamespace(p)) warning(paste0("This vignette needs package `", p, "'; please install"))
}
invisible(suppressMessages(lapply(pkgs, require, character.only = TRUE)))1 liftover
See inst/turbomanin the source, https://github.com/jinghuazhao/pQTLtools/tree/master/inst/turboman, or turboman/ directory in the installed package.
2 ExpressionSet
We start with Bioconductor/Biobase’s ExpressionSet example and finish with a real application.
dataDirectory <- system.file("extdata", package="Biobase")
exprsFile <- file.path(dataDirectory, "exprsData.txt")
exprs <- as.matrix(read.table(exprsFile, header=TRUE, sep="\t", row.names=1, as.is=TRUE))
pDataFile <- file.path(dataDirectory, "pData.txt")
pData <- read.table(pDataFile, row.names=1, header=TRUE, sep="\t")
all(rownames(pData)==colnames(exprs))
metadata <- data.frame(labelDescription=c("Patient gender",
"Case/control status",
"Tumor progress on XYZ scale"),
row.names=c("gender", "type", "score"))
phenoData <- Biobase::AnnotatedDataFrame(data=pData, varMetadata=metadata)
experimentData <- Biobase::MIAME(name="Pierre Fermat",
lab="Francis Galton Lab",
contact="pfermat@lab.not.exist",
title="Smoking-Cancer Experiment",
abstract="An example ExpressionSet",
url="www.lab.not.exist",
other=list(notes="Created from text files"))
exampleSet <- pQTLtools::make_ExpressionSet(exprs,phenoData,experimentData=experimentData,
annotation="hgu95av2")
data(sample.ExpressionSet, package="Biobase")
identical(exampleSet,sample.ExpressionSet)2.1 data.frame
The great benefit is to use the object directly as a data.frame.
lm.result <- Biobase::esApply(exampleSet,1,function(x) lm(score~gender+x))
beta.x <- unlist(lapply(lapply(lm.result,coef),"[",3))
beta.x[1]
#> AFFX-MurIL2_at.x
#> -0.0001907472
lm(score~gender+AFFX.MurIL2_at,data=exampleSet)
#>
#> Call:
#> lm(formula = score ~ gender + AFFX.MurIL2_at, data = exampleSet)
#>
#> Coefficients:
#> (Intercept) genderMale AFFX.MurIL2_at
#> 0.5531725 0.0098932 -0.00019072.2 Composite plots
We wish to examine the distribution of each feature via histogram, scatter and boxplot.
One could resort to esApply() for its simplicity as before
invisible(Biobase::esApply(exampleSet[1:2],1,function(x)
{par(mfrow=c(3,1));boxplot(x);hist(x);plot(x)}
))but it is nicer to add feature name in the title.
par(mfrow=c(1,3))
f <- Biobase::featureNames(exampleSet[1:2])
invisible(sapply(f,function(x) {
d <- t(Biobase::exprs(exampleSet[x]))
fn <- Biobase::featureNames(exampleSet[x])
hist(d,main="",xlab=fn); plot(d, ylab=fn); boxplot(d,ylab=fn)
}
)
)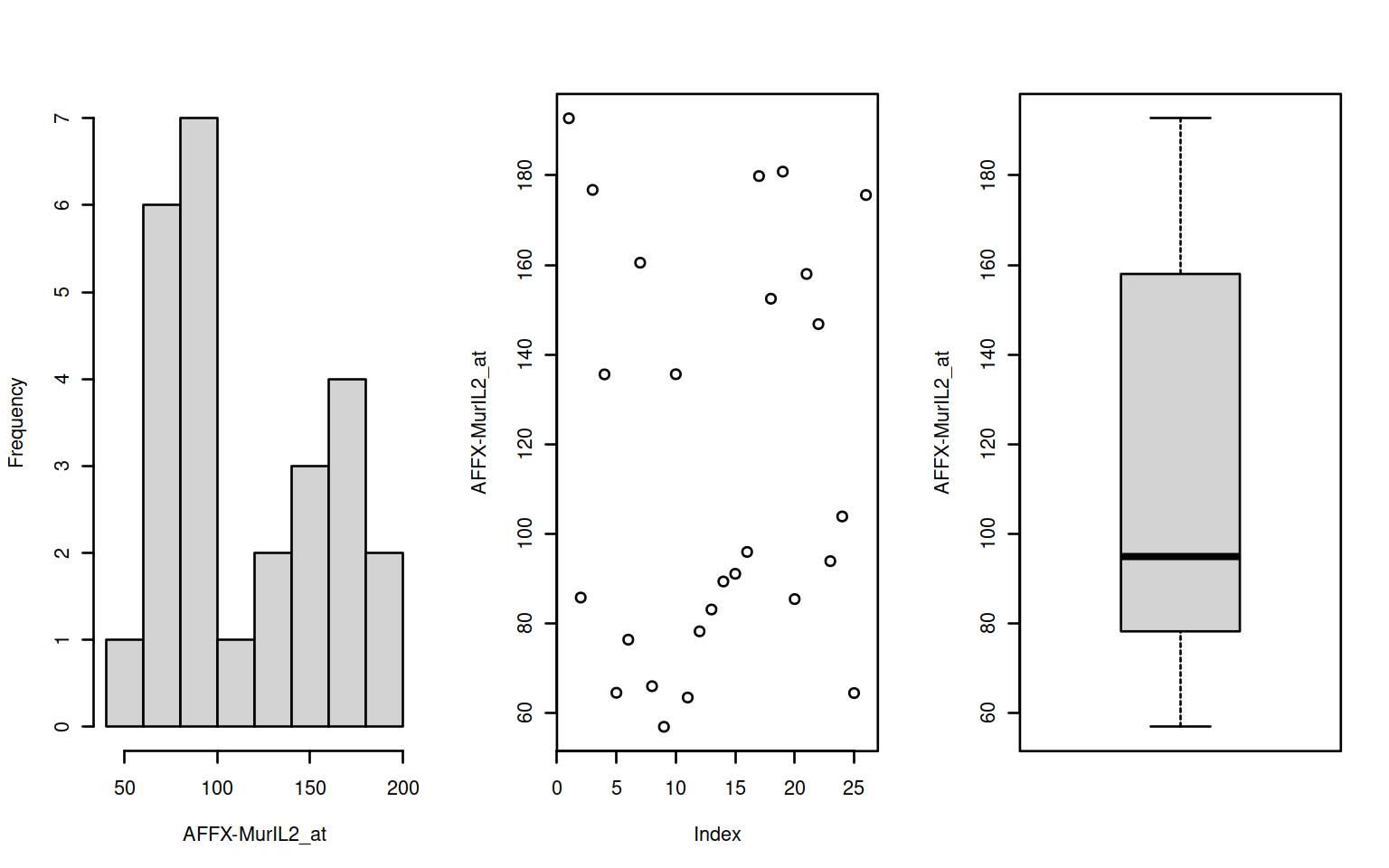
Figure 2.1: Histogram, scatter & boxplots
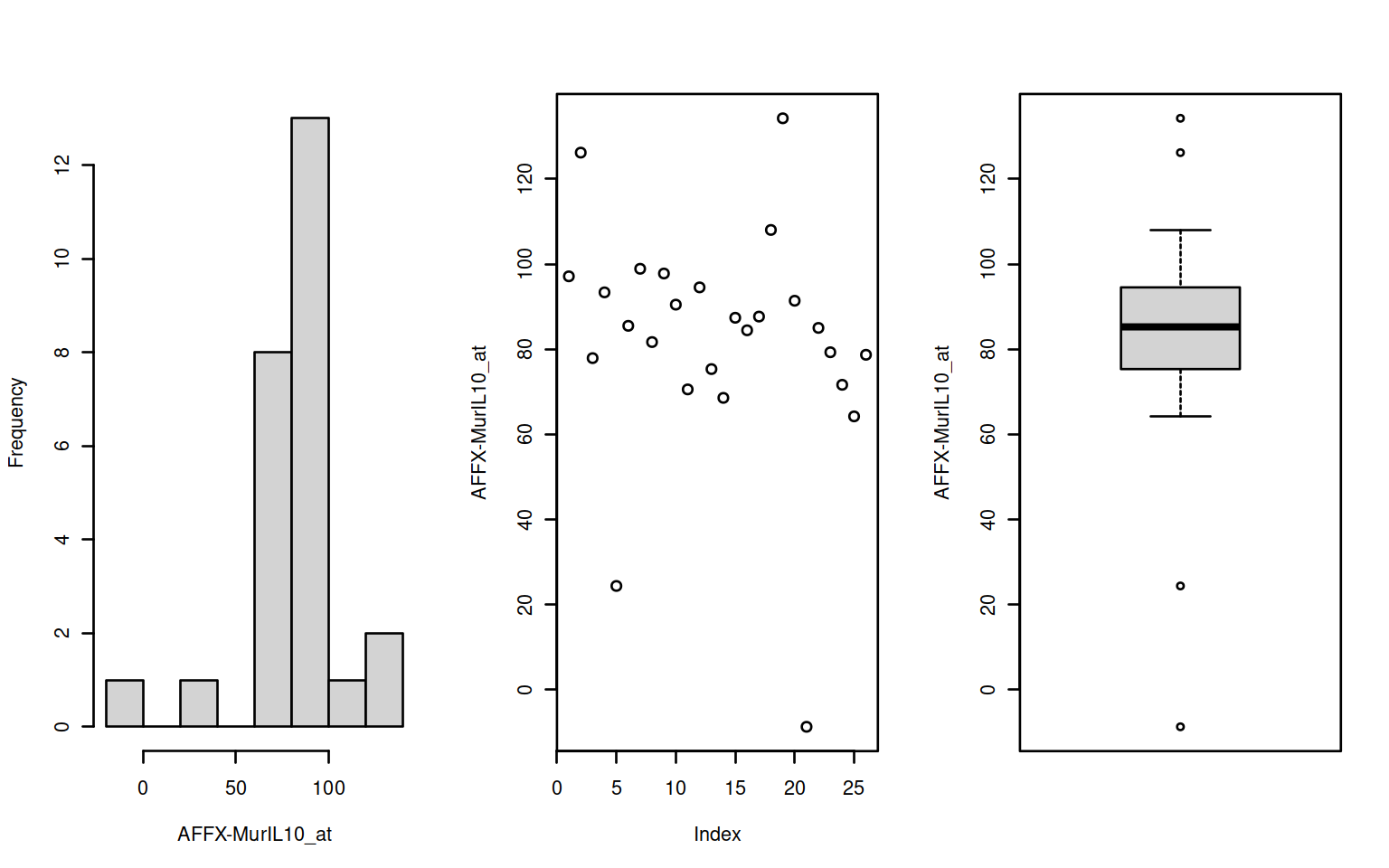
Figure 2.2: Histogram, scatter & boxplots
where the expression set is indexed using feature name(s).
2.4 Clustering
We employ model-based clustering absed on principal compoents to see potential groupings in the data,
pca <- prcomp(na.omit(t(Biobase::exprs(exampleSet))), rank=10, scale=TRUE)
pc1pc2pc3 <- with(pca,x)[,1:3]
mc <- mclust::Mclust(pc1pc2pc3,G=3)
with(mc, {
cols <- c("blue","red", "purple")
s3d <- scatterplot3d::scatterplot3d(with(pca,x[,c(2,1,3)]),
color=cols[classification],
pch=16,
type="h",
main="Plot of the PC1, PC2 and PC3")
s3d.coords <- s3d$xyz.convert(with(pca,x[,c(2,1,3)]))
text(s3d.coords$x,
s3d.coords$y,
cex = 1.2,
col = cols[classification],
labels = row.names(pc1pc2pc3),
pos = 4)
legend("right", legend=levels(as.factor(classification)), col=cols[classification], pch=16)
rgl::open3d(width = 500, height = 500)
rgl::plot3d(with(pca,x[,c(2,1,3)]),cex=1.2,col=cols[classification],size=5)
rgl::text3d(with(pca,x[,c(2,1,3)]),cex=1.2,col=cols[classification],texts=row.names(pc1pc2pc3))
htmlwidgets::saveWidget(rgl::rglwidget(), file = "mcpca3d.html")
})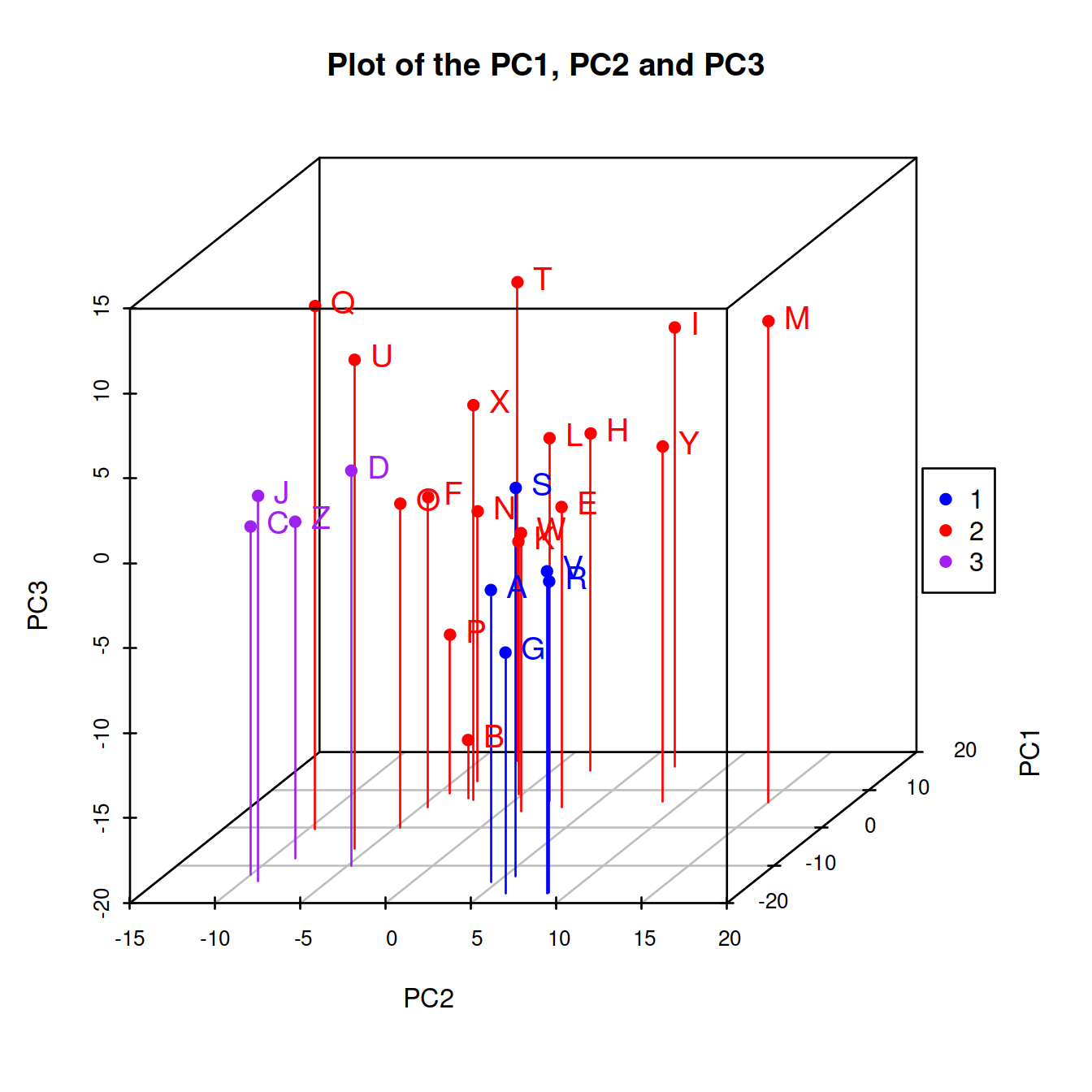
Figure 2.3: Three-group Clustering
#> Warning in par3d(userMatrix = structure(c(1, 0, 0, 0, 0, 0.342020143325668, :
#> parameter "width" cannot be set
#> Warning in par3d(userMatrix = structure(c(1, 0, 0, 0, 0, 0.342020143325668, :
#> parameter "height" cannot be setAn interactive version is also available,
2.5 Data transformation
Suppose we wish use log2 for those greater than zero but set those negative values to be missing.
2.6 Limit of detection (LOD)
We generate a lod.max ~ U[0,1] variable to experiment
Biobase::fData(exampleSet)
#> data frame with 0 columns and 500 rows
Biobase::fData(exampleSet)$lod.max <- apply(Biobase::exprs(exampleSet),1,quantile,runif(nrow(exampleSet)))
lod <- pQTLtools::get.prop.below.LLOD(exampleSet)
x <- dplyr::arrange(Biobase::fData(lod),desc(pc.belowLOD.new))
knitr::kable(head(lod))| AFFX.MurIL2_at | AFFX.MurIL10_at | AFFX.MurIL4_at | AFFX.MurFAS_at | AFFX.BioB.5_at | AFFX.BioB.M_at | gender | type | score | |
|---|---|---|---|---|---|---|---|---|---|
| A | 192.7420 | 97.13700 | 45.81920 | 22.54450 | 96.7875 | 89.0730 | Female | Control | 0.75 |
| B | 85.7533 | 126.19600 | 8.83135 | 3.60093 | 30.4380 | 25.8461 | Male | Case | 0.40 |
| C | 176.7570 | 77.92160 | 33.06320 | 14.68830 | 46.1271 | 57.2033 | Male | Control | 0.73 |
| D | 135.5750 | 93.37130 | 28.70720 | 12.33970 | 70.9319 | 69.9766 | Male | Case | 0.42 |
| E | 64.4939 | 24.39860 | 5.94492 | 36.86630 | 56.1744 | 49.5822 | Female | Case | 0.93 |
| F | 76.3569 | 85.50880 | 28.29250 | 11.25680 | 42.6756 | 26.1262 | Male | Control | 0.22 |
| G | 160.5050 | 98.90860 | 30.96940 | 23.00340 | 86.5156 | 75.0083 | Male | Case | 0.96 |
| H | 65.9631 | 81.69320 | 14.79230 | 16.21340 | 30.7927 | 42.3352 | Male | Case | 0.79 |
| I | 56.9039 | 97.80150 | 14.23990 | 12.03750 | 19.7183 | 41.1207 | Female | Case | 0.37 |
| J | 135.6080 | 90.48380 | 34.48740 | 4.54978 | 46.3520 | 91.5307 | Male | Control | 0.63 |
| K | 63.4432 | 70.57330 | 20.35210 | 8.51782 | 39.1326 | 39.9136 | Male | Case | 0.26 |
| L | 78.2126 | 94.54180 | 14.15540 | 27.28520 | 41.7698 | 49.8397 | Female | Control | 0.36 |
| M | 83.0943 | 75.34550 | 20.62510 | 10.16160 | 80.2197 | 63.4794 | Male | Case | 0.41 |
| N | 89.3372 | 68.58270 | 15.92310 | 20.24880 | 36.4903 | 24.7007 | Male | Case | 0.80 |
| O | 91.0615 | 87.40500 | 20.15790 | 15.78490 | 36.4021 | 47.4641 | Female | Case | 0.10 |
| P | 95.9377 | 84.45810 | 27.81390 | 14.32760 | 35.3054 | 47.3578 | Female | Control | 0.41 |
| Q | 179.8450 | 87.68060 | 32.79110 | 15.94880 | 58.6239 | 58.1331 | Female | Case | 0.16 |
| R | 152.4670 | 108.03200 | 33.52920 | 14.67530 | 114.0620 | 104.1220 | Male | Control | 0.72 |
| S | 180.8340 | 134.26300 | 19.81720 | -7.91911 | 93.4402 | 115.8310 | Male | Case | 0.17 |
| T | 85.4146 | 91.40310 | 20.41900 | 12.88750 | 22.5168 | 58.1224 | Female | Case | 0.74 |
| U | 157.9890 | -8.68811 | 26.87200 | 11.91860 | 48.6462 | 73.4221 | Male | Control | 0.35 |
| V | 146.8000 | 85.02120 | 31.14880 | 12.83240 | 90.2215 | 64.6066 | Female | Control | 0.77 |
| W | 93.8829 | 79.29980 | 22.34200 | 11.13900 | 42.0053 | 40.3068 | Male | Control | 0.27 |
| X | 103.8550 | 71.65520 | 19.01350 | 7.55564 | 57.5738 | 41.8209 | Male | Control | 0.98 |
| Y | 64.4340 | 64.23690 | 12.16860 | 19.98490 | 44.8216 | 46.1087 | Female | Case | 0.94 |
| Z | 175.6150 | 78.70680 | 17.37800 | 8.96849 | 61.7044 | 49.4122 | Female | Case | 0.32 |
plot(x[,2], main="Random quantile cutoff", ylab="<lod%")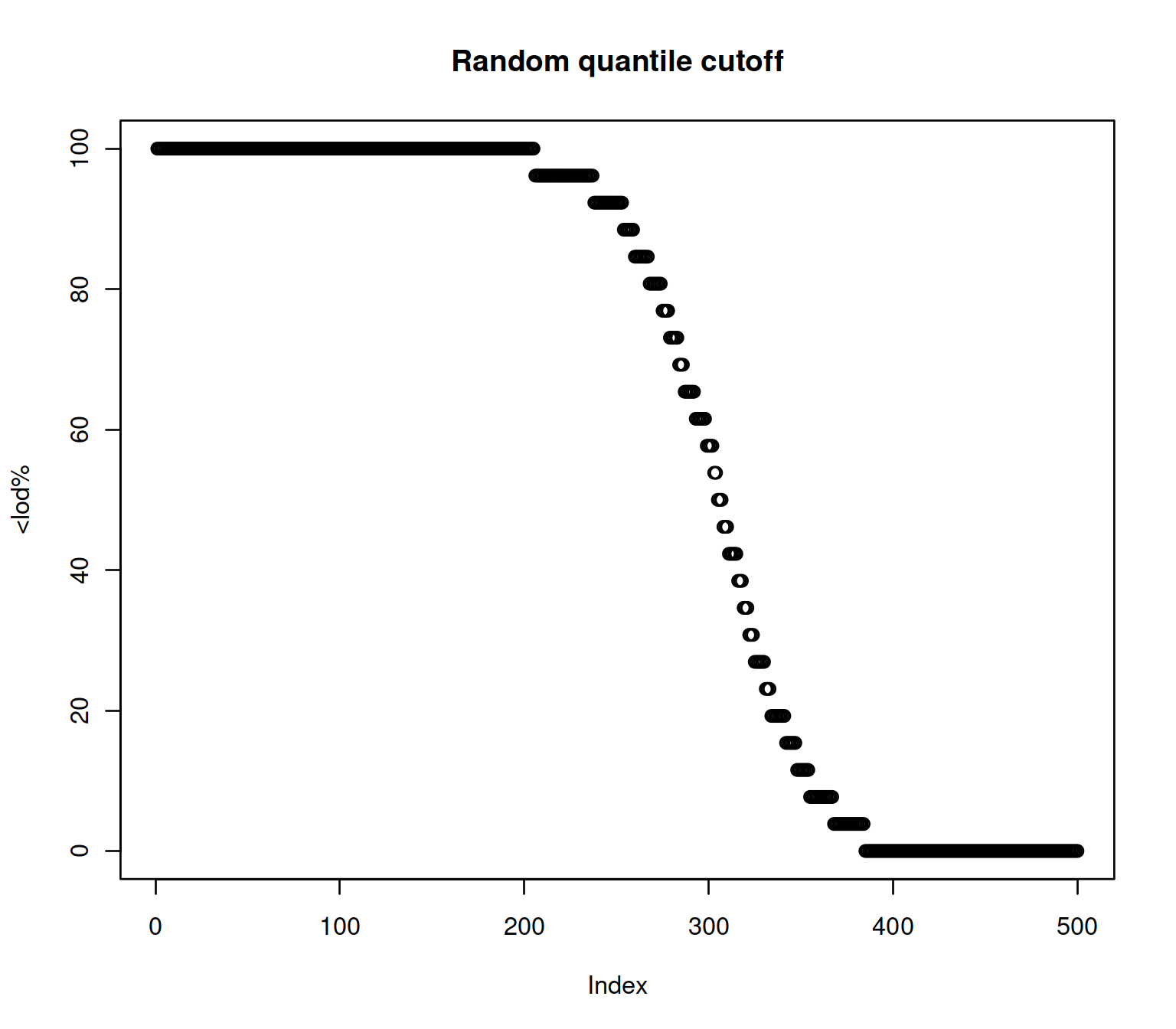
Figure 2.4: LOD based on a random cutoff
The quantity has been shown to have a big impact on protein abundance and therefore pQTL detection as is shown with a real example.
rm(list=ls())
dir <- "~/rds/post_qc_data/interval/phenotype/olink_proteomics/post-qc/"
eset <- readRDS(paste0(dir,"eset.inf1.flag.out.outlier.in.rds"))
x <- pQTLtools::get.prop.below.LLOD(eset)
annot <- Biobase::fData(x)
annot$MissDataProp <- as.numeric(gsub("\\%$", "", annot$MissDataProp))
plot(annot$MissDataProp, annot$pc.belowLOD.new, xlab="% <LLOD in Original",
ylab="% <LLOD in post QC dataset", pch=19)
INF <- Sys.getenv("INF")
np <- read.table(paste(INF, "work", "INF1.merge.nosig", sep="/"), header=FALSE,
col.names = c("prot", "uniprot"))
kable(np, caption="Proteins with no pQTL")
annot$pQTL <- rep(NA, nrow(annot))
no.pQTL.ind <- which(annot$uniprot.id %in% np$uniprot)
annot$pQTL[no.pQTL.ind] <- "red"
annot$pQTL[-no.pQTL.ind] <- "blue"
annot <- annot[order(annot$pc.belowLOD.new, decreasing = TRUE),]
annot <- annot[-grep("^BDNF$", annot$ID),]
saveRDS(annot,file=file.path("~","pQTLtools","tests","annot.RDS"))
annot <- readRDS(file.path(find.package("pQTLtools"),"tests","annot.RDS")) %>%
dplyr::left_join(pQTLdata::inf1[c("prot","target.short","alt_name")],by=c("ID"="prot")) %>%
dplyr::mutate(prot=if_else(is.na(alt_name),target.short,alt_name),order=1:n()) %>%
dplyr::arrange(desc(order))
xtick <- seq(1, nrow(annot))
attach(annot)
par(mar=c(10,5,1,1))
plot(100-pc.belowLOD.new,cex=2,pch=19,col=pQTL,xaxt="n",xlab="",ylab="",cex.axis=0.8)
text(66,16,"IL-17C",offset=0,pos=2,cex=1.5,font=2,srt=0)
arrows(67,16,71,16,lwd=2)
axis(1, at=xtick, labels=prot, lwd.tick=0.5, lwd=0, las=2, hadj=1, cex.axis=0.8)
mtext("% samples above LLOD",side=2,line=2.5,cex=1.2)
mtext("Ordered protein",side=1,line=6.5,cex=1.2,font=1)
legend(x=1,y=25,c("without pQTL","with pQTL"),box.lwd=0,cex=2,col=c("red","blue"),pch=19)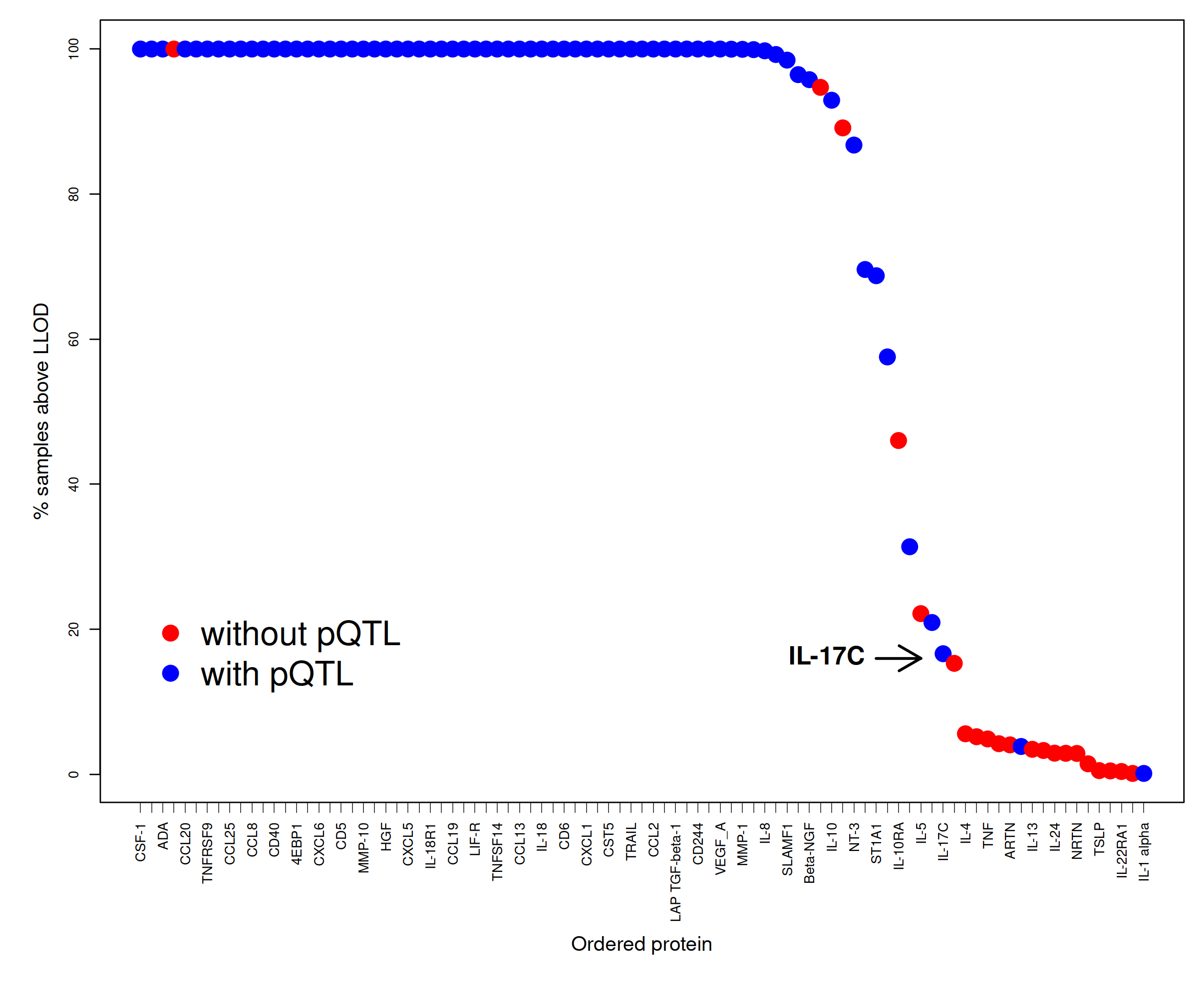
Figure 2.5: LOD in SCALLOP-INF/INTERVAL
| ID | dichot | olink.id | uniprot.id | lod.max | MissDataProp | pc.belowLOD.new | pQTL | target.short | alt_name | prot | order |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CSF.1 | FALSE | 196_CSF-1 | P09603 | 1.02 | 0.02 | 0.0000000 | blue | CSF-1 | NA | CSF-1 | 91 |
| TNFB | FALSE | 195_TNFB | P01374 | 1.10 | 0.04 | 0.0000000 | blue | TNFB | NA | TNFB | 90 |
| ADA | FALSE | 194_ADA | P00813 | 1.80 | 0.04 | 0.0000000 | blue | ADA | NA | ADA | 89 |
| STAMPB | FALSE | 192_STAMPB | O95630 | 1.40 | 0.06 | 0.0000000 | red | STAMPB | NA | STAMPB | 88 |
| CCL20 | FALSE | 190_CCL20 | P78556 | 1.42 | 0.02 | 0.0000000 | blue | CCL20 | NA | CCL20 | 87 |
| TWEAK | FALSE | 189_TWEAK | O43508 | 1.78 | 0.02 | 0.0000000 | blue | TWEAK | NA | TWEAK | 86 |
| TNFRSF9 | FALSE | 187_TNFRSF9 | Q07011 | 1.86 | 0.02 | 0.0000000 | blue | TNFRSF9 | NA | TNFRSF9 | 85 |
| CX3CL1 | FALSE | 186_CX3CL1 | P78423 | 1.76 | 0.02 | 0.0000000 | blue | CX3CL1 | NA | CX3CL1 | 84 |
| CCL25 | FALSE | 185_CCL25 | O15444 | 1.32 | 0.02 | 0.0000000 | blue | CCL25 | NA | CCL25 | 83 |
| CASP.8 | FALSE | 184_CASP-8 | Q14790 | 1.38 | 0.04 | 0.0000000 | blue | CASP-8 | NA | CASP-8 | 82 |
| MCP.2 | FALSE | 183_MCP-2 | P80075 | 2.10 | 0.02 | 0.0000000 | blue | MCP-2 | CCL8 | CCL8 | 81 |
| FGF.19 | FALSE | 179_FGF-19 | O95750 | 1.25 | 0.02 | 0.0000000 | blue | FGF-19 | NA | FGF-19 | 80 |
| CD40 | FALSE | 176_CD40 | P25942 | 1.91 | 0.04 | 0.0000000 | blue | CD40 | NA | CD40 | 79 |
| DNER | FALSE | 174_DNER | Q8NFT8 | 1.63 | 0.02 | 0.0000000 | blue | DNER | NA | DNER | 78 |
| 4E.BP1 | FALSE | 170_4E-BP1 | Q13541 | 1.04 | 0.10 | 0.0000000 | blue | 4EBP1 | NA | 4EBP1 | 77 |
| CXCL10 | FALSE | 169_CXCL10 | P02778 | 2.03 | 0.02 | 0.0000000 | blue | CXCL10 | NA | CXCL10 | 76 |
| CXCL6 | FALSE | 168_CXCL6 | P80162 | 1.80 | 0.02 | 0.0000000 | blue | CXCL6 | CXCL6 | CXCL6 | 75 |
| Flt3L | FALSE | 167_Flt3L | P49771 | 2.12 | 0.02 | 0.0000000 | blue | FIt3L | NA | FIt3L | 74 |
| CD5 | FALSE | 165_CD5 | P06127 | 1.42 | 0.02 | 0.0000000 | blue | CD5 | NA | CD5 | 73 |
| CCL23 | FALSE | 164_CCL23 | P55773 | 2.10 | 0.02 | 0.0000000 | blue | CCL23 | NA | CCL23 | 72 |
| MMP.10 | FALSE | 161_MMP-10 | P09238 | 1.56 | 0.02 | 0.0000000 | blue | MMP-10 | NA | MMP-10 | 71 |
| IL.12B | FALSE | 157_IL-12B | P29460 | 0.85 | 0.02 | 0.0000000 | blue | IL-12B | NA | IL-12B | 70 |
| HGF | FALSE | 156_HGF | P14210 | 1.77 | 0.02 | 0.0000000 | blue | HGF | NA | HGF | 69 |
| TRANCE | FALSE | 155_TRANCE | O14788 | 1.51 | 0.04 | 0.0000000 | blue | TRANCE | NA | TRANCE | 68 |
| CXCL5 | FALSE | 154_CXCL5 | P42830 | 1.92 | 0.02 | 0.0000000 | blue | CXCL5 | NA | CXCL5 | 67 |
| PD.L1 | FALSE | 152_PD-L1 | Q9NZQ7 | 2.14 | 0.04 | 0.0000000 | blue | PD-L1 | NA | PD-L1 | 66 |
| IL.18R1 | FALSE | 151_IL-18R1 | Q13478 | 1.21 | 0.02 | 0.0000000 | blue | IL-18R1 | NA | IL-18R1 | 65 |
| IL.10RB | FALSE | 149_IL-10RB | Q08334 | 1.72 | 0.02 | 0.0000000 | blue | IL10RB | NA | IL10RB | 64 |
| CCL19 | FALSE | 145_CCL19 | Q99731 | 2.13 | 0.02 | 0.0000000 | blue | CCL19 | NA | CCL19 | 63 |
| FGF.21 | FALSE | 144_FGF-21 | Q9NSA1 | 1.14 | 0.02 | 0.0000000 | blue | FGF-21 | NA | FGF-21 | 62 |
| LIF.R | FALSE | 143_LIF-R | P42702 | 2.08 | 0.04 | 0.0000000 | blue | LIF-R | NA | LIF-R | 61 |
| FGF.23 | FALSE | 139_FGF-23 | Q9GZV9 | 0.66 | 0.04 | 0.0000000 | blue | FGF-23 | NA | FGF-23 | 60 |
| TNFSF14 | FALSE | 138_TNFSF14 | O43557 | 1.81 | 0.02 | 0.0000000 | blue | TNFSF14 | NA | TNFSF14 | 59 |
| CCL11 | FALSE | 137_CCL11 | P51671 | 2.09 | 0.02 | 0.0000000 | blue | CCL11 | CCL11 | CCL11 | 58 |
| MCP.4 | FALSE | 136_MCP-4 | Q99616 | 0.56 | 0.04 | 0.0000000 | blue | MCP-4 | CCL13 | CCL13 | 57 |
| TGF.alpha | FALSE | 135_TGF-alpha | P01135 | 0.07 | 0.02 | 0.0000000 | blue | TGF-alpha | NA | TGF-alpha | 56 |
| IL.18 | FALSE | 133_IL-18 | Q14116 | 2.79 | 0.02 | 0.0000000 | blue | IL-18 | NA | IL-18 | 55 |
| SCF | FALSE | 132_SCF | P21583 | 2.30 | 0.02 | 0.0000000 | blue | SCF | NA | SCF | 54 |
| CD6 | FALSE | 131_CD6 | Q8WWJ7 | 1.78 | 0.04 | 0.0000000 | blue | CD6 | NA | CD6 | 53 |
| CCL4 | FALSE | 130_CCL4 | P13236 | 2.15 | 0.02 | 0.0000000 | blue | CCL4 | NA | CCL4 | 52 |
| CXCL1 | FALSE | 128_CXCL1 | P09341 | 3.45 | 0.02 | 0.0000000 | blue | CXCL1 | NA | CXCL1 | 51 |
| OSM | FALSE | 126_OSM | P13725 | 1.32 | 0.06 | 0.0000000 | blue | OSM | NA | OSM | 50 |
| CST5 | FALSE | 123_CST5 | P28325 | 4.34 | 0.04 | 0.0000000 | blue | CST5 | NA | CST5 | 49 |
| CXCL9 | FALSE | 122_CXCL9 | Q07325 | 1.04 | 0.02 | 0.0000000 | blue | CXCL9 | NA | CXCL9 | 48 |
| TRAIL | FALSE | 120_TRAIL | P50591 | 1.22 | 0.02 | 0.0000000 | blue | TRAIL | NA | TRAIL | 47 |
| CXCL11 | FALSE | 117_CXCL11 | O14625 | 1.40 | 0.02 | 0.0000000 | blue | CXCL11 | NA | CXCL11 | 46 |
| MCP.1 | FALSE | 115_MCP-1 | P13500 | 1.90 | 0.02 | 0.0000000 | blue | MCP-1 | CCL2 | CCL2 | 45 |
| uPA | FALSE | 112_uPA | P00749 | 1.75 | 0.02 | 0.0000000 | blue | uPA | NA | uPA | 44 |
| LAP.TGF.beta.1 | FALSE | 111_LAP TGF-beta-1 | P01137 | 0.86 | 0.02 | 0.0000000 | blue | LAP TGF-beta-1 | NA | LAP TGF-beta-1 | 43 |
| OPG | FALSE | 110_OPG | O00300 | 1.81 | 0.02 | 0.0000000 | blue | OPG | NA | OPG | 42 |
| CD244 | FALSE | 108_CD244 | Q9BZW8 | 1.09 | 0.02 | 0.0000000 | blue | CD244 | NA | CD244 | 41 |
| CDCP1 | FALSE | 107_CDCP1 | Q9H5V8 | 0.21 | 0.04 | 0.0000000 | blue | CDCP1 | NA | CDCP1 | 40 |
| VEGF.A | FALSE | 102_VEGF-A | P15692 | 3.15 | 0.02 | 0.0000000 | blue | VEGF_A | NA | VEGF_A | 39 |
| MIP.1.alpha | FALSE | 166_MIP-1 alpha | P10147 | 1.92 | 0.06 | 0.0203998 | blue | MIP-1 alpha | NA | MIP-1 alpha | 38 |
| MMP.1 | FALSE | 142_MMP-1 | P03956 | 1.89 | 0.06 | 0.0407997 | blue | MMP-1 | NA | MMP-1 | 37 |
| SIRT2 | FALSE | 172_SIRT2 | Q8IXJ6 | 1.52 | 1.00 | 0.0815993 | blue | SIRT2 | NA | SIRT2 | 36 |
| IL.8 | FALSE | 101_IL-8 | P10145 | 3.17 | 0.25 | 0.2447980 | blue | IL-8 | NA | IL-8 | 35 |
| IL.7 | FALSE | 109_IL-7 | P13232 | 1.39 | 1.43 | 0.7547940 | blue | IL-7 | NA | IL-7 | 34 |
| SLAMF1 | FALSE | 134_SLAMF1 | Q13291 | 1.55 | 2.13 | 1.5299878 | blue | SLAMF1 | NA | SLAMF1 | 33 |
| EN.RAGE | FALSE | 175_EN-RAGE | P80511 | 1.57 | 3.28 | 3.5291718 | blue | EN-RAGE | NA | EN-RAGE | 32 |
| Beta.NGF | FALSE | 153_Beta-NGF | P01138 | 1.23 | 3.96 | 4.2227662 | blue | Beta-NGF | NA | Beta-NGF | 31 |
| CCL28 | FALSE | 173_CCL28 | Q9NRJ3 | 0.92 | 5.49 | 5.2835577 | red | CCL28 | NA | CCL28 | 30 |
| IL.10 | FALSE | 162_IL-10 | P22301 | 1.50 | 7.71 | 7.0583435 | blue | IL-10 | NA | IL-10 | 29 |
| AXIN1 | FALSE | 118_AXIN1 | O15169 | 1.30 | 13.04 | 10.8731130 | red | AXIN1 | NA | AXIN1 | 28 |
| NT.3 | FALSE | 188_NT-3 | P20783 | 0.85 | 12.77 | 13.2394941 | blue | NT-3 | NA | NT-3 | 27 |
| FGF.5 | FALSE | 141_FGF-5 | Q8NF90 | 1.00 | 31.71 | 30.3957568 | blue | FGF-5 | NA | FGF-5 | 26 |
| ST1A1 | FALSE | 191_ST1A1 | P50225 | 1.34 | 32.68 | 31.2525500 | blue | ST1A1 | NA | ST1A1 | 25 |
| GDNF | FALSE | 106_GDNF | P39905 | 1.13 | 43.26 | 42.4520604 | blue | hGDNF | NA | hGDNF | 24 |
| IL.10RA | FALSE | 140_IL-10RA | Q13651 | 1.14 | 53.30 | 53.9779682 | red | IL-10RA | NA | IL-10RA | 23 |
| IL.6 | FALSE | 113_IL-6 | P05231 | 1.97 | 69.06 | 68.6250510 | blue | IL-6 | NA | IL-6 | 22 |
| IL.5 | FALSE | 193_IL-5 | P05113 | 1.55 | 78.21 | 77.8457772 | red | IL-5 | NA | IL-5 | 21 |
| MCP.3 | FALSE | 105_MCP-3 | P80098 | 1.31 | 79.30 | 79.0697674 | blue | MCP-3 | CCL7 | CCL7 | 20 |
| IL.17C | FALSE | 114_IL-17C | Q9P0M4 | 1.28 | 83.07 | 83.3741330 | blue | IL-17C | NA | IL-17C | 19 |
| IL.17A | FALSE | 116_IL-17A | Q16552 | 0.97 | 84.73 | 84.7001224 | red | IL-17A | NA | IL-17A | 18 |
| IL.4 | FALSE | 180_IL-4 | P05112 | 1.11 | 93.91 | 94.4104447 | red | IL-4 | NA | IL-4 | 17 |
| LIF | FALSE | 181_LIF | P15018 | 1.45 | 94.90 | 94.8184415 | red | LIF | NA | LIF | 16 |
| TNF | FALSE | 163_TNF | P01375 | 1.19 | 95.22 | 95.1244390 | red | TNF | NA | TNF | 15 |
| IL.20RA | FALSE | 121_IL-20RA | Q9UHF4 | 0.79 | 95.16 | 95.7772338 | red | IL-20RA | NA | IL-20RA | 14 |
| ARTN | FALSE | 160_ARTN | Q5T4W7 | 0.73 | 95.84 | 95.9404325 | red | ARTN | NA | ARTN | 13 |
| IL.15RA | FALSE | 148_IL-15RA | Q13261 | 1.18 | 96.10 | 96.1648307 | blue | IL-15RA | NA | IL-15RA | 12 |
| IL.13 | FALSE | 159_IL-13 | P35225 | 1.71 | 96.51 | 96.5524276 | red | IL-13 | NA | IL-13 | 11 |
| IL.20 | FALSE | 171_IL-20 | Q9NYY1 | 1.30 | 96.72 | 96.7156263 | red | IL-20 | NA | IL-20 | 10 |
| IL.24 | FALSE | 158_IL-24 | Q13007 | 1.95 | 97.13 | 97.0828233 | red | IL-24 | NA | IL-24 | 9 |
| IL.2RB | FALSE | 124_IL-2RB | P14784 | 1.47 | 97.11 | 97.1032232 | red | IL-2RB | NA | IL-2RB | 8 |
| NRTN | FALSE | 182_NRTN | Q99748 | 1.39 | 97.11 | 97.1236230 | red | NRTN | NA | NRTN | 7 |
| IL.33 | FALSE | 177_IL-33 | O95760 | 1.17 | 98.54 | 98.5516116 | red | IL-33 | NA | IL-33 | 6 |
| TSLP | FALSE | 129_TSLP | Q969D9 | 1.80 | 99.49 | 99.4900041 | red | TSLP | NA | TSLP | 5 |
| IFN.gamma | FALSE | 178_IFN-gamma | P01579 | 1.40 | 99.53 | 99.5308038 | red | IFN-gamma | NA | IFN-gamma | 4 |
| IL.22.RA1 | FALSE | 150_IL-22 RA1 | Q8N6P7 | 1.79 | 99.61 | 99.6124031 | red | IL-22RA1 | NA | IL-22RA1 | 3 |
| IL.2 | FALSE | 127_IL-2 | P60568 | 0.93 | 99.86 | 99.8776010 | red | IL-2 | NA | IL-2 | 2 |
| IL.1.alpha | FALSE | 125_IL-1 alpha | P01583 | 4.76 | 99.88 | 99.8776010 | blue | IL-1 alpha | NA | IL-1 alpha | 1 |
2.7 maEndtoEnd
Web: https://bioconductor.org/packages/release/workflows/html/maEndToEnd.html.
Examples can be found on PCA, heatmap, normalisation, linear models, and enrichment analysis from this Bioconductor package.
3 SummarizedExperiment
This is a more modern construct. Based on the documentation example, ranged summarized experiment (rse) and imputation are shown below.
set.seed(123)
nrows <- 20
ncols <- 4
counts <- matrix(runif(nrows * ncols, 1, 1e4), nrows)
missing_indices <- sample(length(counts), size = 5, replace = FALSE)
counts[missing_indices] <- NA
rowRanges <- GenomicRanges::GRanges(rep(c("chr1", "chr2"), c(1, 3) * nrows / 4),
IRanges::IRanges(floor(runif(nrows, 1e5, 1e6)), width=ncols),
strand=sample(c("+", "-"), nrows, TRUE),
feature_id=sprintf("ID%03d", 1:nrows))
colData <- S4Vectors::DataFrame(Treatment=rep(c("ChIP", "Input"), ncols/2),
row.names=LETTERS[1:ncols])
rse <- SummarizedExperiment::SummarizedExperiment(assays=S4Vectors::SimpleList(counts=counts),
rowRanges=rowRanges, colData=colData)
SummarizedExperiment::assay(rse)
#> A B C D
#> [1,] 2876.4876 8895.5036 1428.8574 6651.486831
#> [2,] 7883.2630 6928.3413 4146.0488 949.311769
#> [3,] 4090.3602 6405.4276 4137.8295 3840.312408
#> [4,] 8830.2910 9942.7035 3689.0857 2744.562062
#> [5,] 9404.7324 6557.4023 1525.2950 8146.585749
#> [6,] NA 7085.5962 1388.9218 4485.714898
#> [7,] 5281.5268 5441.1162 2331.1080 8100.833466
#> [8,] 8924.2980 5941.8261 4660.1585 8124.082706
#> [9,] 5514.7987 2892.3082 2660.4604 7943.628869
#> [10,] 4566.6907 1471.9894 NA 4398.877044
#> [11,] 9568.3766 NA 459.2658 7544.997111
#> [12,] 4533.8882 9023.0882 4422.5585 NA
#> [13,] 6776.0288 6907.3621 7989.4495 7102.113831
#> [14,] 5726.7614 7954.8787 1219.8707 7.247108
#> [15,] 1030.1439 247.1122 5609.9189 4753.690424
#> [16,] 8998.3499 4778.4819 2066.1074 2201.968733
#> [17,] 2461.6313 7584.8369 1276.1890 3798.785561
#> [18,] 421.5533 2164.8630 7533.3253 6128.097262
#> [19,] 3279.8793 NA 8950.5585 3518.627295
#> [20,] 9545.0820 2317.0262 3745.2533 1112.243108
imputed <- MsCoreUtils::impute_knn(as.matrix(SummarizedExperiment::assay(rse)),2)
imputed_counts <- MsCoreUtils::impute_RF(as.matrix(SummarizedExperiment::assay(rse)),2)
imputed-imputed_counts
#> A B C D
#> [1,] 0.000 0.0000 0.000 0.000
#> [2,] 0.000 0.0000 0.000 0.000
#> [3,] 0.000 0.0000 0.000 0.000
#> [4,] 0.000 0.0000 0.000 0.000
#> [5,] 0.000 0.0000 0.000 0.000
#> [6,] 2559.339 0.0000 0.000 0.000
#> [7,] 0.000 0.0000 0.000 0.000
#> [8,] 0.000 0.0000 0.000 0.000
#> [9,] 0.000 0.0000 0.000 0.000
#> [10,] 0.000 0.0000 1791.909 0.000
#> [11,] 0.000 -709.2786 0.000 0.000
#> [12,] 0.000 0.0000 0.000 2467.614
#> [13,] 0.000 0.0000 0.000 0.000
#> [14,] 0.000 0.0000 0.000 0.000
#> [15,] 0.000 0.0000 0.000 0.000
#> [16,] 0.000 0.0000 0.000 0.000
#> [17,] 0.000 0.0000 0.000 0.000
#> [18,] 0.000 0.0000 0.000 0.000
#> [19,] 0.000 1298.3456 0.000 0.000
#> [20,] 0.000 0.0000 0.000 0.000
SummarizedExperiment::assays(rse) <- S4Vectors::SimpleList(counts=imputed_counts)
SummarizedExperiment::assay(rse)
#> A B C D
#> [1,] 2876.4876 8895.5036 1428.8574 6651.486831
#> [2,] 7883.2630 6928.3413 4146.0488 949.311769
#> [3,] 4090.3602 6405.4276 4137.8295 3840.312408
#> [4,] 8830.2910 9942.7035 3689.0857 2744.562062
#> [5,] 9404.7324 6557.4023 1525.2950 8146.585749
#> [6,] 4439.2947 7085.5962 1388.9218 4485.714898
#> [7,] 5281.5268 5441.1162 2331.1080 8100.833466
#> [8,] 8924.2980 5941.8261 4660.1585 8124.082706
#> [9,] 5514.7987 2892.3082 2660.4604 7943.628869
#> [10,] 4566.6907 1471.9894 4812.3872 4398.877044
#> [11,] 9568.3766 5545.7565 459.2658 7544.997111
#> [12,] 4533.8882 9023.0882 4422.5585 4611.837102
#> [13,] 6776.0288 6907.3621 7989.4495 7102.113831
#> [14,] 5726.7614 7954.8787 1219.8707 7.247108
#> [15,] 1030.1439 247.1122 5609.9189 4753.690424
#> [16,] 8998.3499 4778.4819 2066.1074 2201.968733
#> [17,] 2461.6313 7584.8369 1276.1890 3798.785561
#> [18,] 421.5533 2164.8630 7533.3253 6128.097262
#> [19,] 3279.8793 4566.2289 8950.5585 3518.627295
#> [20,] 9545.0820 2317.0262 3745.2533 1112.243108
SummarizedExperiment::assays(rse) <- S4Vectors::endoapply(SummarizedExperiment::assays(rse), asinh)
SummarizedExperiment::assay(rse)
#> A B C D
#> [1,] 8.657472 9.786448 7.957778 9.495743
#> [2,] 9.665644 9.536523 9.023058 7.548885
#> [3,] 9.009536 9.458048 9.021074 8.946456
#> [4,] 9.779090 9.897741 8.906281 8.610524
#> [5,] 9.842115 9.481497 8.023090 9.698501
#> [6,] 9.091398 9.558966 7.929430 9.101800
#> [7,] 9.265118 9.294887 8.447246 9.692869
#> [8,] 9.789680 9.382919 9.139952 9.695735
#> [9,] 9.308338 8.662957 8.579402 9.673273
#> [10,] 9.119691 7.987517 9.172096 9.082252
#> [11,] 9.859366 9.313936 6.822778 9.621787
#> [12,] 9.112482 9.800689 9.087621 9.129529
#> [13,] 9.514294 9.533490 9.679024 9.561295
#> [14,] 9.346053 9.674688 7.799647 2.678476
#> [15,] 7.630601 6.202994 9.325439 9.159824
#> [16,] 9.797944 9.165025 8.326569 8.390254
#> [17,] 8.501727 9.627054 7.844781 8.935584
#> [18,] 6.737095 8.373260 9.620239 9.413787
#> [19,] 8.788709 9.119590 9.792618 8.858973
#> [20,] 9.856929 8.441187 8.921392 7.707281
SummarizedExperiment::rowRanges(rse)
#> GRanges object with 20 ranges and 1 metadata column:
#> seqnames ranges strand | feature_id
#> <Rle> <IRanges> <Rle> | <character>
#> [1] chr1 409164-409167 - | ID001
#> [2] chr1 691082-691085 + | ID002
#> [3] chr1 388335-388338 + | ID003
#> [4] chr1 268922-268925 - | ID004
#> [5] chr1 804064-804067 - | ID005
#> ... ... ... ... . ...
#> [16] chr2 647861-647864 + | ID016
#> [17] chr2 469620-469623 + | ID017
#> [18] chr2 232385-232388 + | ID018
#> [19] chr2 941769-941772 - | ID019
#> [20] chr2 371106-371109 + | ID020
#> -------
#> seqinfo: 2 sequences from an unspecified genome; no seqlengths
SummarizedExperiment::rowData(rse)
#> DataFrame with 20 rows and 1 column
#> feature_id
#> <character>
#> 1 ID001
#> 2 ID002
#> 3 ID003
#> 4 ID004
#> 5 ID005
#> ... ...
#> 16 ID016
#> 17 ID017
#> 18 ID018
#> 19 ID019
#> 20 ID020
SummarizedExperiment::colData(rse)
#> DataFrame with 4 rows and 1 column
#> Treatment
#> <character>
#> A ChIP
#> B Input
#> C ChIP
#> D Input4 Normalisation
4.1 ComBat
This is the documentation example, based on Bioconductor 3.14.
data(bladderdata, package="bladderbatch")
edat <- bladderEset[1:50]
pheno <- Biobase::pData(edat)
batch <- pheno$batch
table(batch)
#> batch
#> 1 2 3 4 5
#> 11 18 4 5 19
quantro::matboxplot(edat,batch,cex.axis=0.6,notch=TRUE,pch=19,ylab="Expression")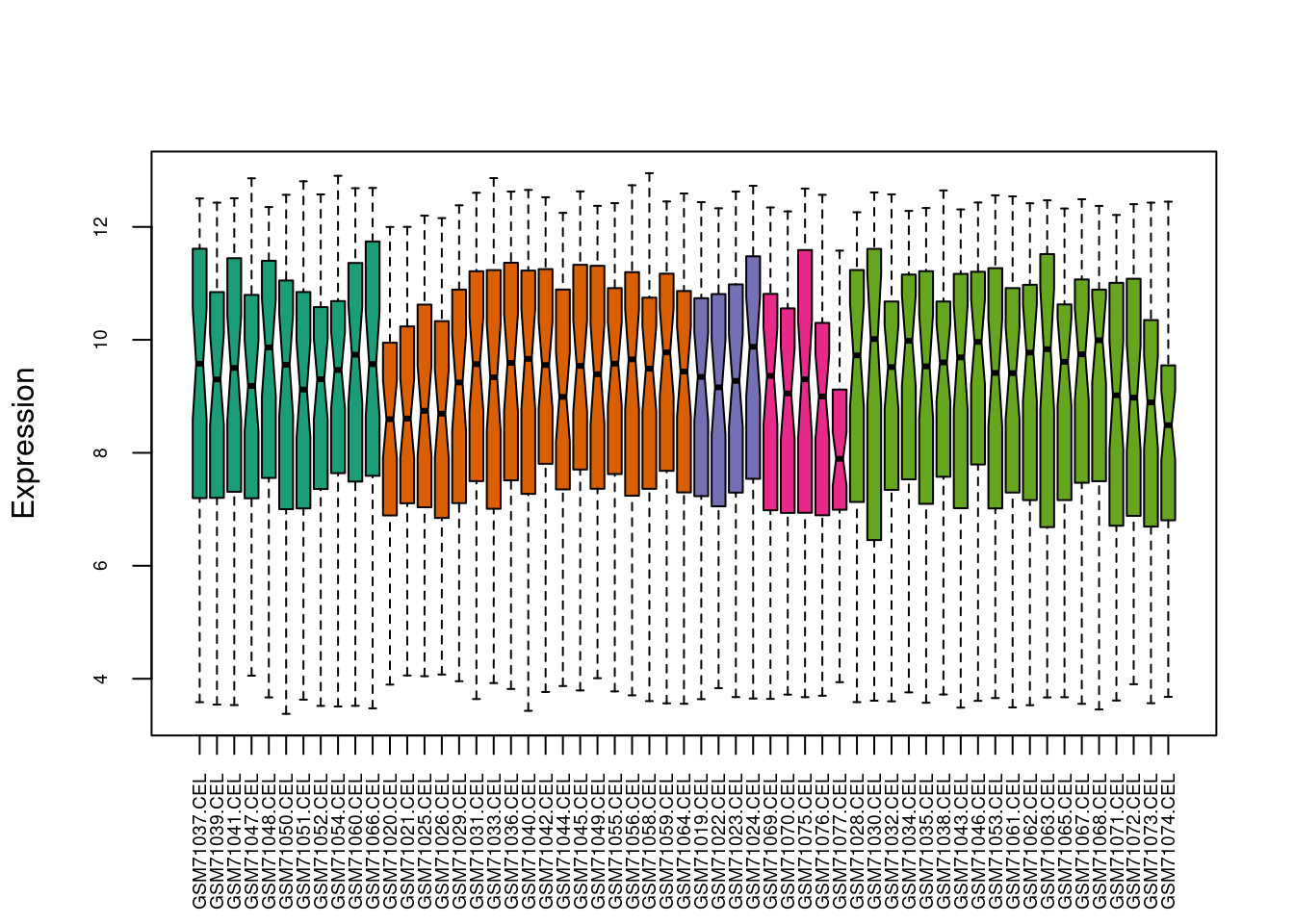
Figure 4.1: ComBat example
quantro::matdensity(edat,batch,xlab=" ",ylab="density")
legend("topleft",legend=1:5,col=1:5,lty=1)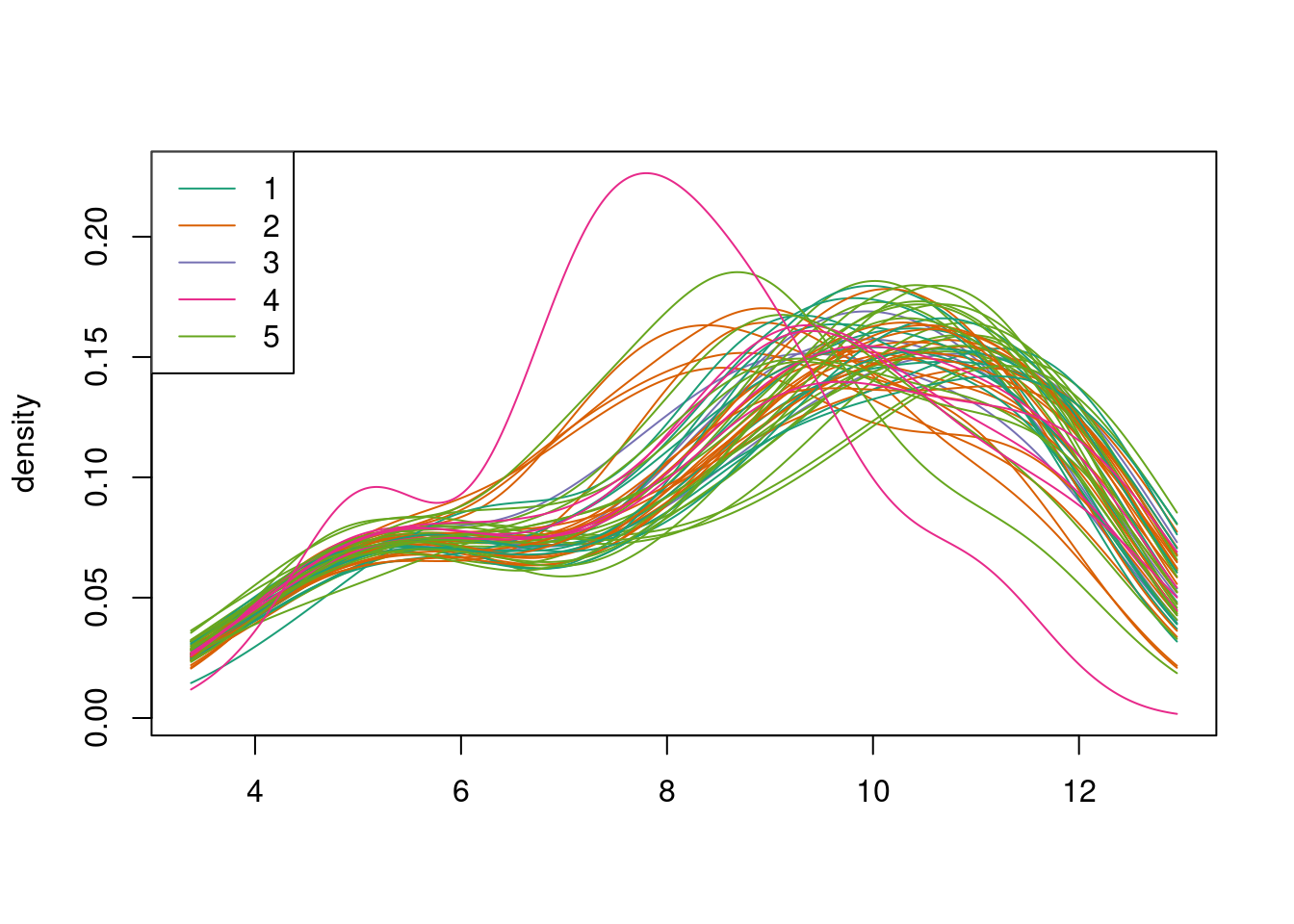
Figure 4.2: ComBat example
# 1. parametric adjustment
combat_edata1 <- sva::ComBat(dat=edat, batch=batch, par.prior=TRUE, prior.plots=TRUE)
#> Found5batches
#> Adjusting for0covariate(s) or covariate level(s)
#> Standardizing Data across genes
#> Fitting L/S model and finding priors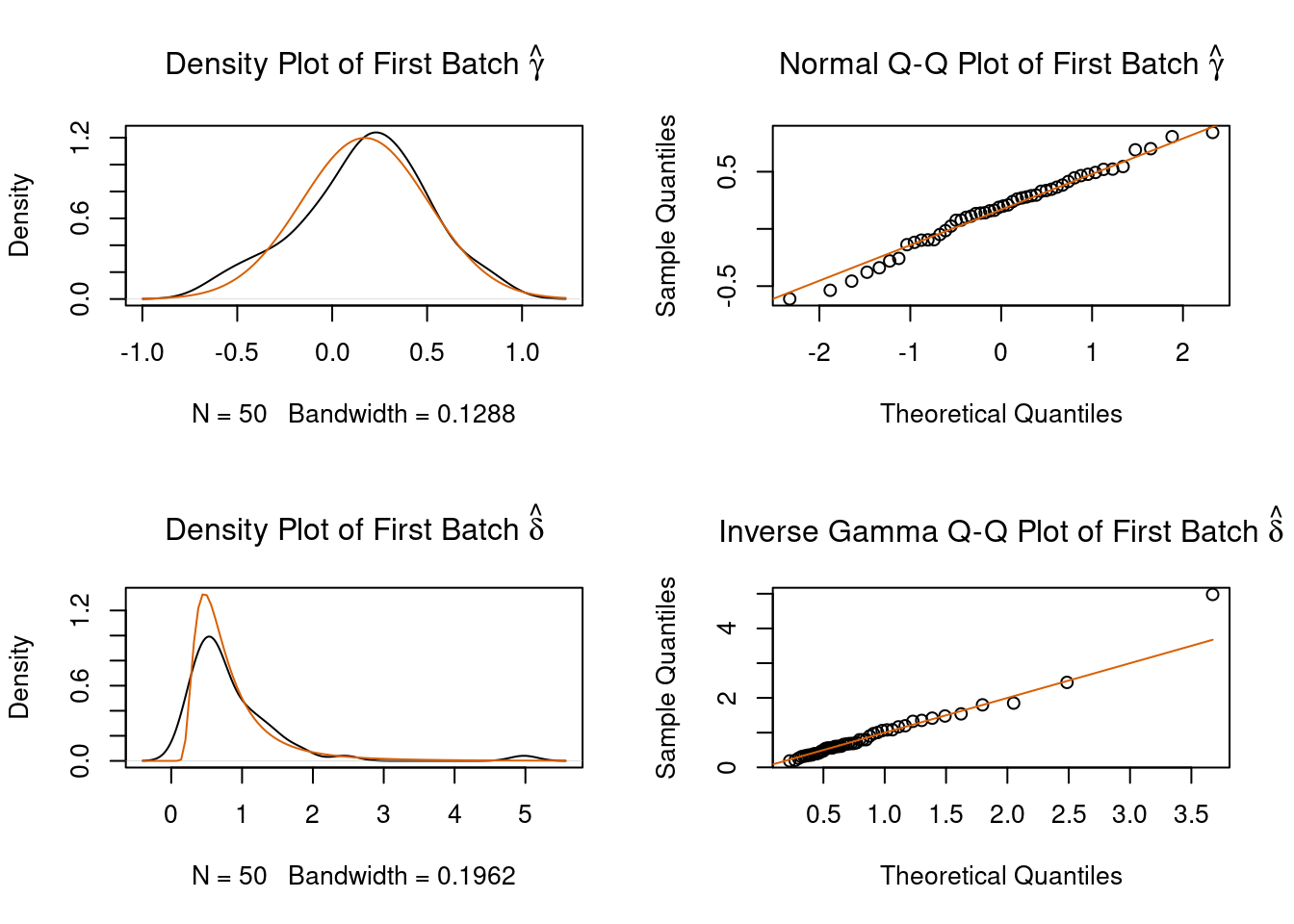
Figure 4.3: ComBat example
#> Finding parametric adjustments
#> Adjusting the Data
# 2. non-parametric adjustment, mean-only version
combat_edata2 <- sva::ComBat(dat=edat, batch=batch, par.prior=FALSE, mean.only=TRUE)
#> Using the 'mean only' version of ComBat
#> Found5batches
#> Adjusting for0covariate(s) or covariate level(s)
#> Standardizing Data across genes
#> Fitting L/S model and finding priors
#> Finding nonparametric adjustments
#> Adjusting the Data
# 3. reference-batch version, with covariates
mod <- model.matrix(~as.factor(cancer), data=pheno)
combat_edata3 <- sva::ComBat(dat=edat, batch=batch, mod=mod, par.prior=TRUE, ref.batch=3, prior.plots=TRUE)
#> Using batch =3as a reference batch (this batch won't change)
#> Found5batches
#> Adjusting for2covariate(s) or covariate level(s)
#> Standardizing Data across genes
#> Fitting L/S model and finding priors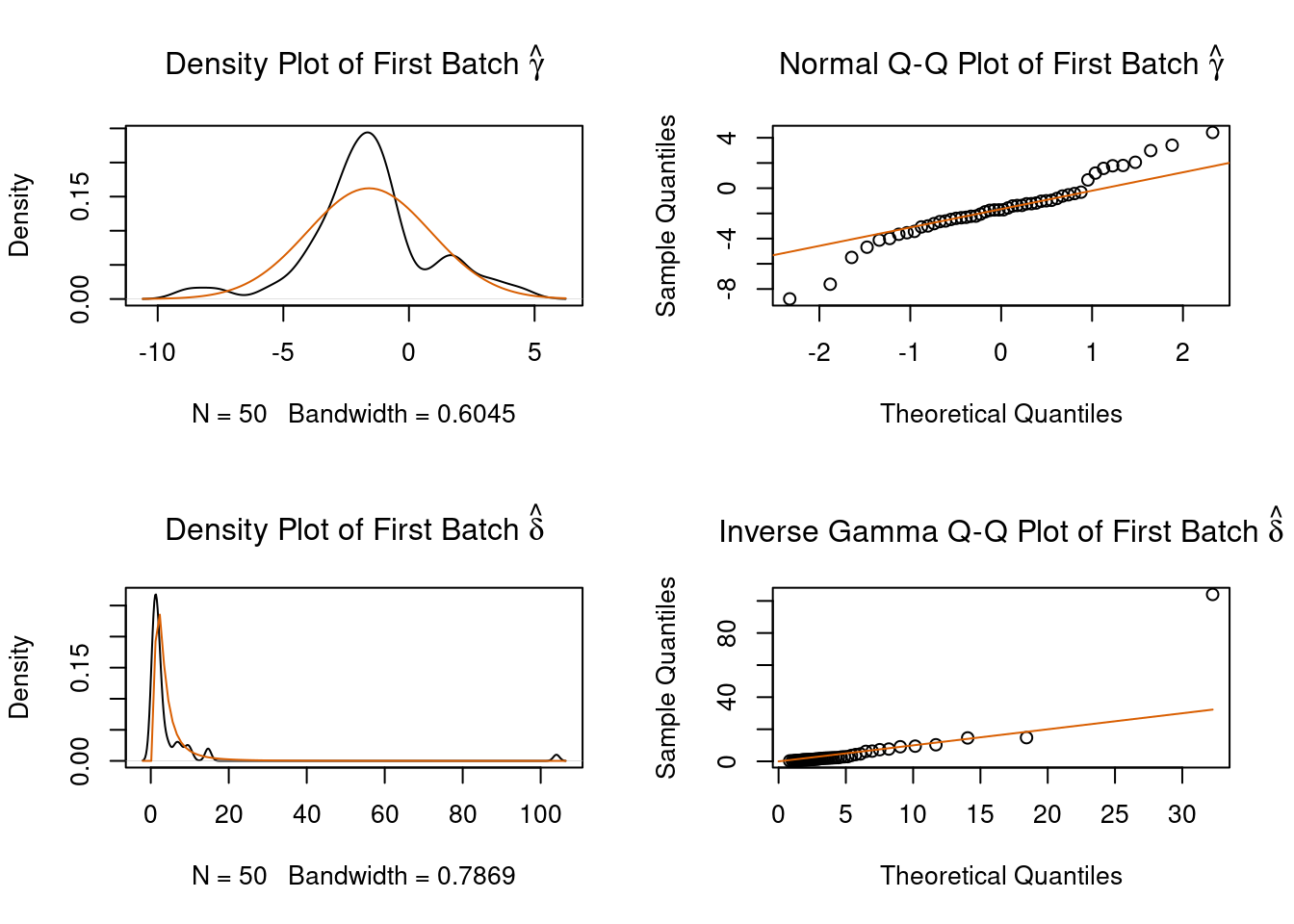
Figure 4.4: ComBat example
#> Finding parametric adjustments
#> Adjusting the Data4.2 quantro
This is also adapted from the package vignette but with FlowSorted.DLPFC.450k in place of FlowSorted.
data(FlowSorted.DLPFC.450k,package="FlowSorted.DLPFC.450k")
p <- getBeta(FlowSorted.DLPFC.450k,offset=100)
pd <- Biobase::pData(FlowSorted.DLPFC.450k)
quantro::matboxplot(p, groupFactor = pd$CellType, xaxt = "n", main = "Beta Values", pch=19)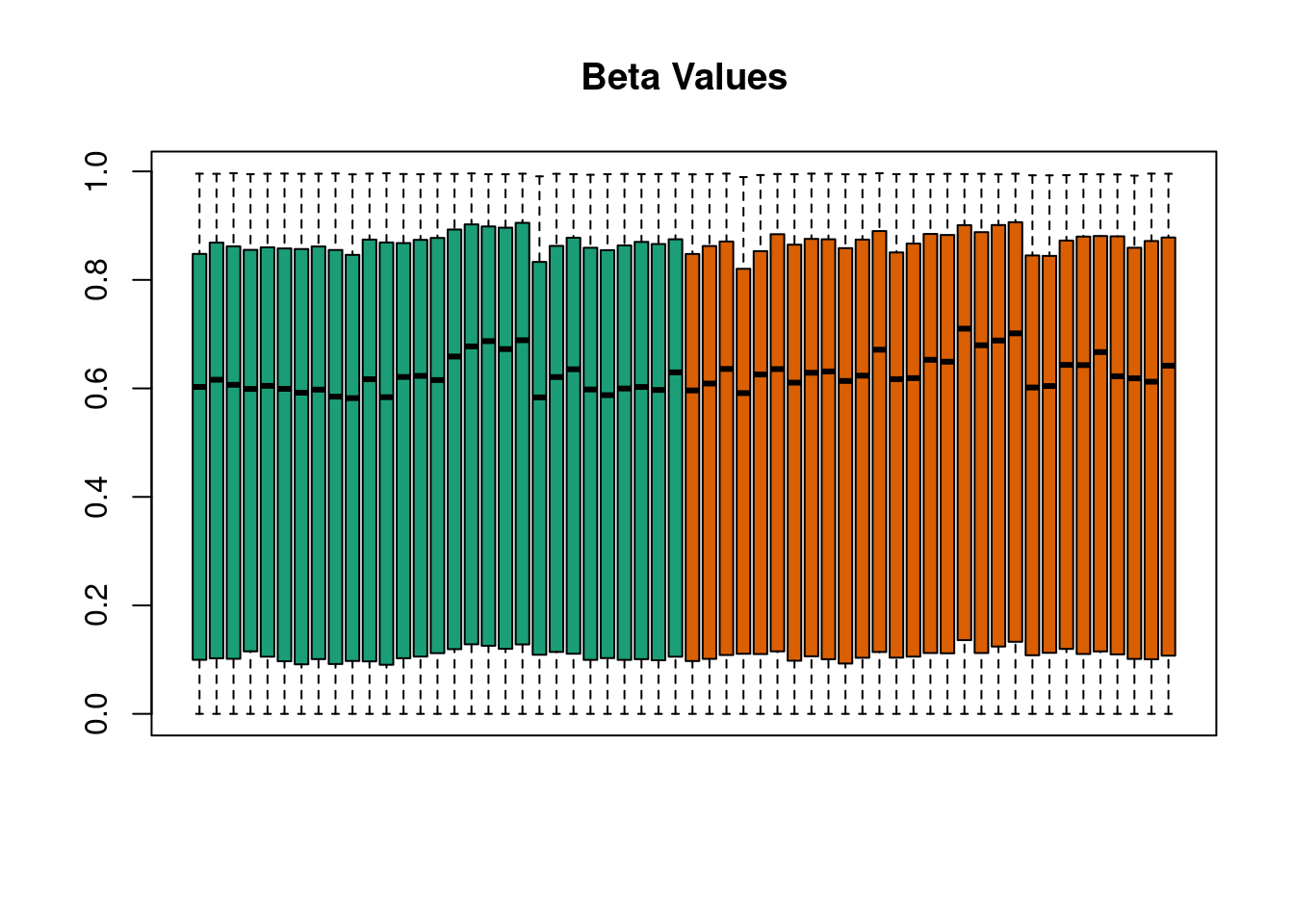
Figure 4.5: quantro example
quantro::matdensity(p, groupFactor = pd$CellType, xlab = " ", ylab = "density",
main = "Beta Values", brewer.n = 8, brewer.name = "Dark2")
legend('top', c("NeuN_neg", "NeuN_pos"), col = c(1, 2), lty = 1, lwd = 3)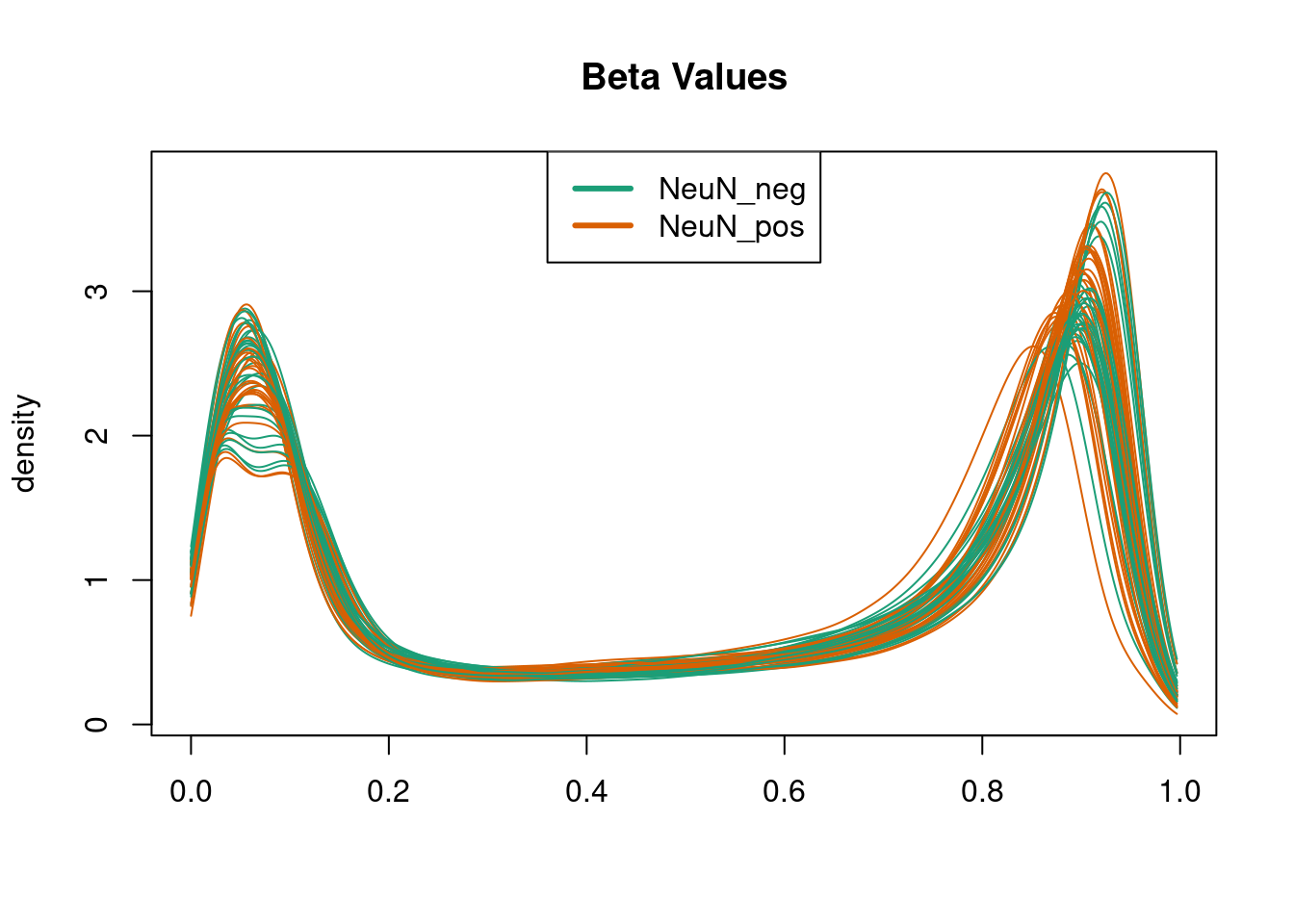
Figure 4.6: quantro example
qtest <- quantro::quantro(object = p, groupFactor = pd$CellType)
#> [quantro] Average medians of the distributions are
#> not equal across groups.
#> [quantro] Calculating the quantro test statistic.
#> [quantro] No permutation testing performed.
#> Use B > 0 for permutation testing.
if (FALSE)
{
doParallel::registerDoParallel(cores=10)
qtestPerm <- quantro::quantro(p, groupFactor = pd$CellType, B = 1000)
quantro::quantroPlot(qtestPerm)
}5 Outlier detection in RNA-Seq
The following is adapted from package OUTRIDER, noting its plotQQ() has issues
ctsFile <- system.file('extdata', 'KremerNBaderSmall.tsv', package='OUTRIDER')
ctsTable <- read.table(ctsFile, check.names=FALSE)
ods <- OUTRIDER::OutriderDataSet(countData=ctsTable)
ods <- OUTRIDER::filterExpression(ods, minCounts=TRUE, filterGenes=TRUE)
#> 229 genes did not pass the filter due to zero counts. This is 22.9% of the genes.
ods <- OUTRIDER::OUTRIDER(ods)
#> Tue Mar 25 10:41:03 2025: SizeFactor estimation ...
#> Tue Mar 25 10:41:03 2025: Controlling for confounders ...
#> Using estimated q with: 23
#> Tue Mar 25 10:41:03 2025: Using the autoencoder implementation for controlling.
#> [1] "Tue Mar 25 10:41:07 2025: Initial PCA loss: 4.73997327486604"
#> [1] "Tue Mar 25 10:41:11 2025: Iteration: 1 loss: 4.19416269506454"
#> [1] "Tue Mar 25 10:41:14 2025: Iteration: 2 loss: 4.17550752619036"
#> [1] "Tue Mar 25 10:41:16 2025: Iteration: 3 loss: 4.16639365666912"
#> [1] "Tue Mar 25 10:41:18 2025: Iteration: 4 loss: 4.16142359470334"
#> [1] "Tue Mar 25 10:41:20 2025: Iteration: 5 loss: 4.15785341106832"
#> [1] "Tue Mar 25 10:41:22 2025: Iteration: 6 loss: 4.15533343090454"
#> [1] "Tue Mar 25 10:41:23 2025: Iteration: 7 loss: 4.15339892434562"
#> [1] "Tue Mar 25 10:41:25 2025: Iteration: 8 loss: 4.15175378925737"
#> [1] "Tue Mar 25 10:41:26 2025: Iteration: 9 loss: 4.15069201289976"
#> [1] "Tue Mar 25 10:41:27 2025: Iteration: 10 loss: 4.1501222741797"
#> [1] "Tue Mar 25 10:41:29 2025: Iteration: 11 loss: 4.14904801948777"
#> [1] "Tue Mar 25 10:41:30 2025: Iteration: 12 loss: 4.14805452270911"
#> [1] "Tue Mar 25 10:41:31 2025: Iteration: 13 loss: 4.14796461892655"
#> [1] "Tue Mar 25 10:41:32 2025: Iteration: 14 loss: 4.14722109314569"
#> [1] "Tue Mar 25 10:41:33 2025: Iteration: 15 loss: 4.14696284053289"
#> Time difference of 25.25854 secs
#> [1] "Tue Mar 25 10:41:33 2025: 15 Final nb-AE loss: 4.14696284053289"
#> Tue Mar 25 10:41:33 2025: Used the autoencoder implementation for controlling.
#> Tue Mar 25 10:41:33 2025: P-value calculation ...
#> Tue Mar 25 10:41:37 2025: Zscore calculation ...
res <- OUTRIDER::results(ods)
knitr::kable(res,caption="A check list of outliers")| geneID | sampleID | pValue | padjust | zScore | l2fc | rawcounts | meanRawcounts | normcounts | meanCorrected | theta | aberrant | AberrantBySample | AberrantByGene | padj_rank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATAD3C | MUC1360 | 0.0e+00 | 0.0000001 | 5.28 | 1.88 | 948 | 82.29 | 248.14 | 67.33 | 16.48 | TRUE | 1 | 1 | 1 |
| NBPF15 | MUC1351 | 0.0e+00 | 0.0000035 | 5.80 | 0.77 | 7591 | 4224.88 | 7028.35 | 4121.14 | 112.78 | TRUE | 2 | 1 | 1 |
| MSTO1 | MUC1367 | 0.0e+00 | 0.0000177 | -6.26 | -0.81 | 761 | 1327.87 | 727.74 | 1276.02 | 153.26 | TRUE | 1 | 1 | 1 |
| HDAC1 | MUC1350 | 0.0e+00 | 0.0001133 | -5.88 | -0.78 | 2215 | 3805.56 | 2128.19 | 3649.04 | 135.60 | TRUE | 1 | 1 | 1 |
| DCAF6 | MUC1374 | 1.0e-07 | 0.0004182 | -5.67 | -0.61 | 2348 | 4869.53 | 3085.40 | 4724.25 | 196.31 | TRUE | 1 | 1 | 1 |
| NBPF16 | MUC1351 | 2.0e-07 | 0.0006791 | 4.83 | 0.67 | 4014 | 2459.90 | 3822.18 | 2402.36 | 107.85 | TRUE | 2 | 1 | 2 |
| FAM102B | MUC1363 | 1.2e-06 | 0.0067682 | -5.37 | -1.29 | 455 | 1138.75 | 440.62 | 1076.81 | 41.98 | TRUE | 1 | 1 | 1 |
| LOC100288142 | MUC1361 | 3.0e-06 | 0.0167927 | 4.25 | 0.85 | 637 | 356.12 | 622.40 | 345.76 | 57.44 | TRUE | 1 | 1 | 1 |
| TARDBP | MUC0486 | 7.3e-06 | 0.0407384 | -4.56 | -0.32 | 5911 | 5780.34 | 4449.25 | 5565.44 | 464.81 | TRUE | 1 | 1 | 1 |
| ZMPSTE24 | MUC1370 | 7.7e-06 | 0.0426990 | 4.28 | 0.38 | 6180 | 4026.77 | 5088.19 | 3905.38 | 273.05 | TRUE | 1 | 1 | 1 |
6 Differential expression
ex <- DESeq2::makeExampleDESeqDataSet(m=4)
dds <- DESeq2::DESeq(ex)
#> estimating size factors
#> estimating dispersions
#> gene-wise dispersion estimates
#> mean-dispersion relationship
#> final dispersion estimates
#> fitting model and testing
res <- DESeq2::results(dds, contrast=c("condition","B","A"))
rld <- DESeq2::rlogTransformation(ex, blind=TRUE)
dat <- DESeq2::plotPCA(rld, intgroup=c("condition"),returnData=TRUE)
#> using ntop=500 top features by variance
percentVar <- round(100 * attr(dat,"percentVar"))
ggplot2::ggplot(dat, ggplot2::aes(PC1, PC2, color=condition, shape=condition)) +
ggplot2::geom_point(size=3) +
ggplot2::xlab(paste0("PC1:",percentVar[1],"% variance")) +
ggplot2::ylab(paste0("PC2:",percentVar[2],"% variance"))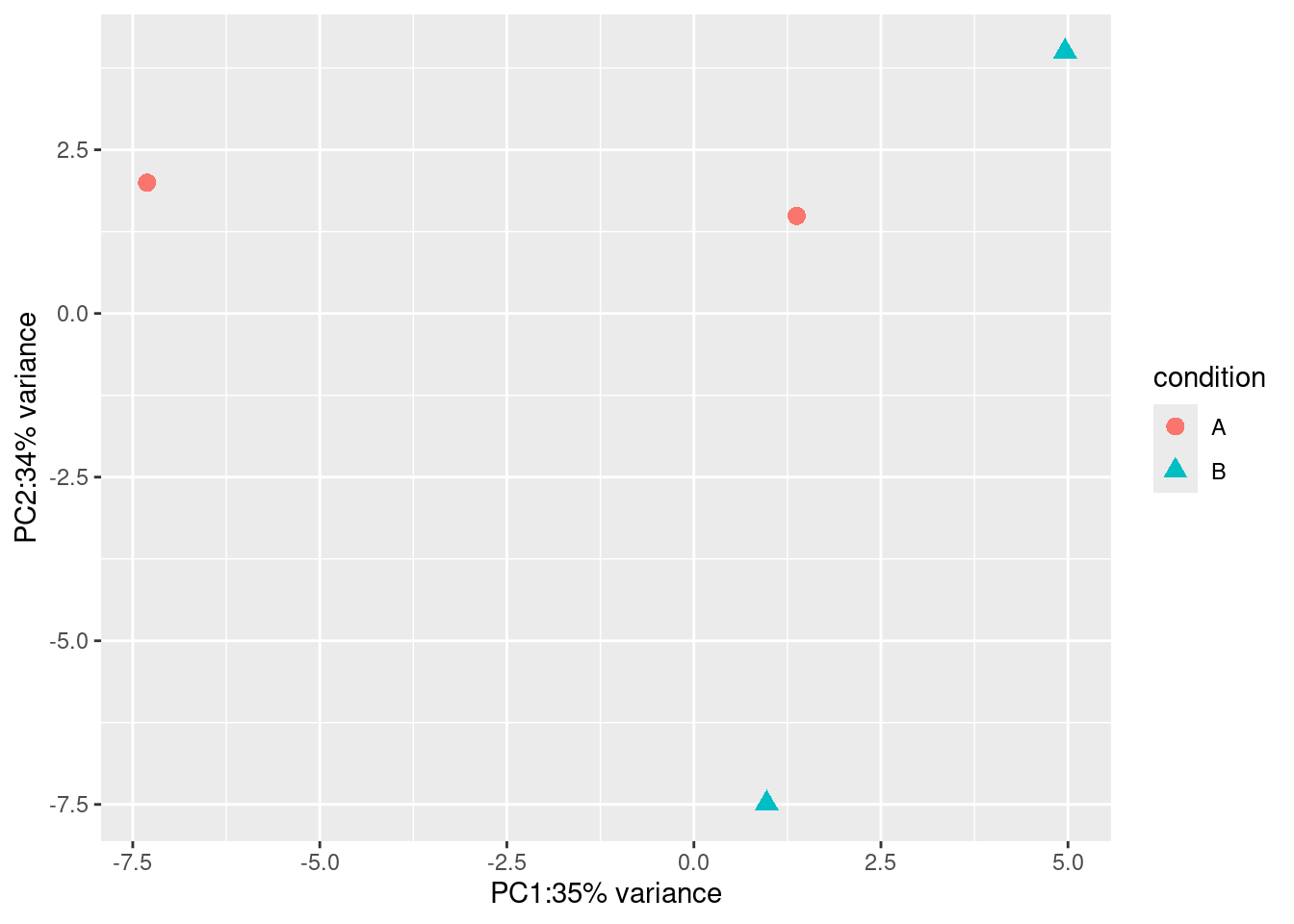
Figure 6.1: DESeq2 example
ex$condition <- relevel(ex$condition, ref="B")
dds2 <- DESeq2::DESeq(dds)
#> using pre-existing size factors
#> estimating dispersions
#> found already estimated dispersions, replacing these
#> gene-wise dispersion estimates
#> mean-dispersion relationship
#> final dispersion estimates
#> fitting model and testing
res <- DESeq2::results(dds2)
knitr::kable(head(as.data.frame(res)))| baseMean | log2FoldChange | lfcSE | stat | pvalue | padj | |
|---|---|---|---|---|---|---|
| gene1 | 8.884414 | 0.2449020 | 1.3370638 | 0.1831640 | 0.8546693 | 0.9896044 |
| gene2 | 18.554877 | -0.9545927 | 1.0945708 | -0.8721161 | 0.3831451 | 0.9896044 |
| gene3 | 10.398828 | -0.0389598 | 1.2211301 | -0.0319047 | 0.9745481 | 0.9987191 |
| gene4 | 13.075617 | 1.1475896 | 1.1265255 | 1.0186983 | 0.3083462 | 0.9896044 |
| gene5 | 10.629715 | -0.3700499 | 1.2156482 | -0.3044054 | 0.7608190 | 0.9896044 |
| gene6 | 22.092276 | -1.4125098 | 0.9883968 | -1.4290918 | 0.1529778 | 0.9896044 |
See the package in action from a snakemake workflow1.
7 Gene co-expression and network analysis
A simple network is furnished with the GeneNet documentation example,
## A random network with 40 nodes
# it contains 780=40*39/2 edges of which 5 percent (=39) are non-zero
true.pcor <- GeneNet::ggm.simulate.pcor(40)
# A data set with 40 observations
m.sim <- GeneNet::ggm.simulate.data(40, true.pcor)
# A simple estimate of partial correlations
estimated.pcor <- corpcor::cor2pcor( cor(m.sim) )
# A comparison of estimated and true values
sum((true.pcor-estimated.pcor)^2)
#> [1] 346.6915
# A slightly better estimate ...
estimated.pcor.2 <- GeneNet::ggm.estimate.pcor(m.sim)
#> Estimating optimal shrinkage intensity lambda (correlation matrix): 0.2942
sum((true.pcor-estimated.pcor.2)^2)
#> [1] 11.19183
## ecoli data
data(ecoli, package="GeneNet")
# partial correlation matrix
inferred.pcor <- GeneNet::ggm.estimate.pcor(ecoli)
#> Estimating optimal shrinkage intensity lambda (correlation matrix): 0.1804
# p-values, q-values and posterior probabilities for each potential edge
test.results <- GeneNet::network.test.edges(inferred.pcor)
#> Estimate (local) false discovery rates (partial correlations):
#> Step 1... determine cutoff point
#> Step 2... estimate parameters of null distribution and eta0
#> Step 3... compute p-values and estimate empirical PDF/CDF
#> Step 4... compute q-values and local fdr
#> Step 5... prepare for plotting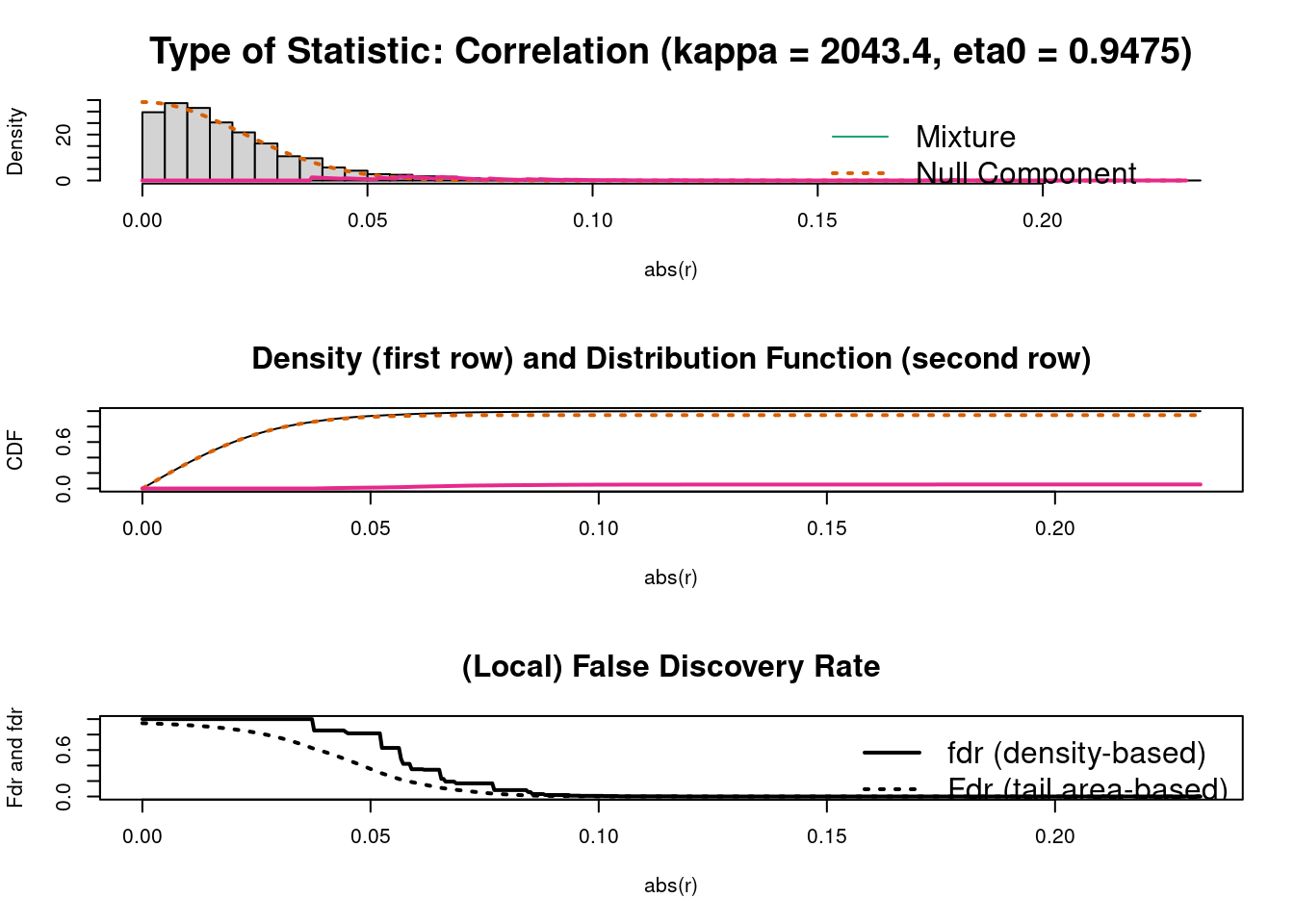
Figure 7.1: GeneNet example
# best 20 edges (strongest correlation)
test.results[1:20,]
#> pcor node1 node2 pval qval prob
#> 1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
#> 2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
#> 3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
#> 4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
#> 5 -0.13418892 29 86 1.120812e-09 1.093997e-06 0.9999516
#> 6 0.12594697 21 72 1.103836e-08 8.978563e-06 0.9998400
#> 7 0.11956105 28 86 5.890924e-08 3.853590e-05 0.9998400
#> 8 -0.11723897 26 80 1.060526e-07 5.816172e-05 0.9998400
#> 9 -0.11711625 72 89 1.093655e-07 5.930499e-05 0.9972804
#> 10 0.10658013 20 21 1.366610e-06 5.925275e-04 0.9972804
#> 11 0.10589778 21 73 1.596859e-06 6.678429e-04 0.9972804
#> 12 0.10478689 20 91 2.053403e-06 8.024425e-04 0.9972804
#> 13 0.10420836 7 52 2.338382e-06 8.778605e-04 0.9944557
#> 14 0.10236077 87 95 3.525186e-06 1.224964e-03 0.9944557
#> 15 0.10113550 27 95 4.610444e-06 1.500047e-03 0.9920084
#> 16 0.09928954 21 51 6.868357e-06 2.046549e-03 0.9920084
#> 17 0.09791914 21 88 9.192373e-06 2.520616e-03 0.9920084
#> 18 0.09719685 18 95 1.070232e-05 2.790102e-03 0.9920084
#> 19 0.09621791 28 90 1.313007e-05 3.171817e-03 0.9920084
#> 20 0.09619099 12 80 1.320374e-05 3.182526e-03 0.9920084
# network containing edges with prob > 0.9 (i.e. local fdr < 0.1)
net <- GeneNet::extract.network(test.results, cutoff.ggm=0.9)
#>
#> Significant edges: 65
#> Corresponding to 1.26 % of possible edges
net
#> pcor node1 node2 pval qval prob
#> 1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
#> 2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
#> 3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
#> 4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
#> 5 -0.13418892 29 86 1.120812e-09 1.093997e-06 0.9999516
#> 6 0.12594697 21 72 1.103836e-08 8.978563e-06 0.9998400
#> 7 0.11956105 28 86 5.890924e-08 3.853590e-05 0.9998400
#> 8 -0.11723897 26 80 1.060526e-07 5.816172e-05 0.9998400
#> 9 -0.11711625 72 89 1.093655e-07 5.930499e-05 0.9972804
#> 10 0.10658013 20 21 1.366610e-06 5.925275e-04 0.9972804
#> 11 0.10589778 21 73 1.596859e-06 6.678429e-04 0.9972804
#> 12 0.10478689 20 91 2.053403e-06 8.024425e-04 0.9972804
#> 13 0.10420836 7 52 2.338382e-06 8.778605e-04 0.9944557
#> 14 0.10236077 87 95 3.525186e-06 1.224964e-03 0.9944557
#> 15 0.10113550 27 95 4.610444e-06 1.500047e-03 0.9920084
#> 16 0.09928954 21 51 6.868357e-06 2.046549e-03 0.9920084
#> 17 0.09791914 21 88 9.192373e-06 2.520616e-03 0.9920084
#> 18 0.09719685 18 95 1.070232e-05 2.790102e-03 0.9920084
#> 19 0.09621791 28 90 1.313007e-05 3.171817e-03 0.9920084
#> 20 0.09619099 12 80 1.320374e-05 3.182526e-03 0.9920084
#> 21 0.09576091 89 95 1.443542e-05 3.354777e-03 0.9891317
#> 22 0.09473210 7 51 1.784126e-05 3.864825e-03 0.9891317
#> 23 -0.09386896 53 58 2.127622e-05 4.313590e-03 0.9891317
#> 24 -0.09366615 29 83 2.217013e-05 4.421099e-03 0.9891317
#> 25 -0.09341148 21 89 2.334321e-05 4.556947e-03 0.9810727
#> 26 -0.09156391 49 93 3.380043e-05 5.955972e-03 0.9810727
#> 27 -0.09150710 80 90 3.418363e-05 6.002083e-03 0.9810727
#> 28 0.09101505 7 53 3.767966e-05 6.408102e-03 0.9810727
#> 29 0.09050688 21 84 4.164471e-05 6.838782e-03 0.9810727
#> 30 0.08965490 72 73 4.919365e-05 7.581866e-03 0.9810727
#> 31 -0.08934025 29 99 5.229604e-05 7.861416e-03 0.9810727
#> 32 -0.08906819 9 95 5.512708e-05 8.104759e-03 0.9810727
#> 33 0.08888345 2 49 5.713144e-05 8.270673e-03 0.9810727
#> 34 0.08850681 86 90 6.143363e-05 8.610161e-03 0.9810727
#> 35 0.08805868 17 53 6.695170e-05 9.015175e-03 0.9810727
#> 36 0.08790809 28 48 6.890884e-05 9.151291e-03 0.9810727
#> 37 0.08783471 33 58 6.988211e-05 9.217597e-03 0.9682377
#> 38 -0.08705796 7 49 8.101244e-05 1.021362e-02 0.9682377
#> 39 0.08645033 20 46 9.086547e-05 1.102466e-02 0.9682377
#> 40 0.08609950 48 86 9.705862e-05 1.150392e-02 0.9682377
#> 41 0.08598769 21 52 9.911458e-05 1.165816e-02 0.9682377
#> 42 0.08555275 32 95 1.075099e-04 1.226435e-02 0.9682377
#> 43 0.08548231 17 51 1.089311e-04 1.236337e-02 0.9424721
#> 44 0.08470370 80 83 1.258659e-04 1.382356e-02 0.9424721
#> 45 0.08442510 80 82 1.325062e-04 1.437068e-02 0.9174573
#> 46 0.08271606 80 93 1.810275e-04 1.845632e-02 0.9174573
#> 47 0.08235175 46 91 1.933329e-04 1.941579e-02 0.9174573
#> 48 0.08217787 25 95 1.994788e-04 1.988432e-02 0.9174573
#> 49 -0.08170331 29 87 2.171999e-04 2.119715e-02 0.9174573
#> 50 0.08123632 19 29 2.360716e-04 2.253606e-02 0.9174573
#> 51 0.08101702 51 84 2.454547e-04 2.318024e-02 0.9174573
#> 52 0.08030748 16 93 2.782643e-04 2.532796e-02 0.9174573
#> 53 0.08006503 28 52 2.903870e-04 2.608271e-02 0.9174573
#> 54 -0.07941656 41 80 3.252833e-04 2.814824e-02 0.9174573
#> 55 0.07941410 54 89 3.254229e-04 2.815620e-02 0.9174573
#> 56 -0.07934653 28 80 3.292784e-04 2.837511e-02 0.9174573
#> 57 0.07916783 29 92 3.396802e-04 2.895702e-02 0.9174573
#> 58 -0.07866905 17 86 3.703635e-04 3.060293e-02 0.9174573
#> 59 0.07827749 16 29 3.962446e-04 3.191462e-02 0.9174573
#> 60 -0.07808262 73 89 4.097452e-04 3.257290e-02 0.9174573
#> 61 0.07766261 52 67 4.403165e-04 3.400207e-02 0.9174573
#> 62 0.07762917 25 87 4.428396e-04 3.411637e-02 0.9174573
#> 63 -0.07739378 9 93 4.609872e-04 3.492295e-02 0.9174573
#> 64 0.07738885 31 80 4.613747e-04 3.493988e-02 0.9174573
#> 65 -0.07718681 80 94 4.775136e-04 3.563444e-02 0.9174573
# significant based on FDR cutoff Q=0.05?
num.significant.1 <- sum(test.results$qval <= 0.05)
test.results[1:num.significant.1,]
#> pcor node1 node2 pval qval prob
#> 1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
#> 2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
#> 3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
#> 4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
#> 5 -0.13418892 29 86 1.120812e-09 1.093997e-06 0.9999516
#> 6 0.12594697 21 72 1.103836e-08 8.978563e-06 0.9998400
#> 7 0.11956105 28 86 5.890924e-08 3.853590e-05 0.9998400
#> 8 -0.11723897 26 80 1.060526e-07 5.816172e-05 0.9998400
#> 9 -0.11711625 72 89 1.093655e-07 5.930499e-05 0.9972804
#> 10 0.10658013 20 21 1.366610e-06 5.925275e-04 0.9972804
#> 11 0.10589778 21 73 1.596859e-06 6.678429e-04 0.9972804
#> 12 0.10478689 20 91 2.053403e-06 8.024425e-04 0.9972804
#> 13 0.10420836 7 52 2.338382e-06 8.778605e-04 0.9944557
#> 14 0.10236077 87 95 3.525186e-06 1.224964e-03 0.9944557
#> 15 0.10113550 27 95 4.610444e-06 1.500047e-03 0.9920084
#> 16 0.09928954 21 51 6.868357e-06 2.046549e-03 0.9920084
#> 17 0.09791914 21 88 9.192373e-06 2.520616e-03 0.9920084
#> 18 0.09719685 18 95 1.070232e-05 2.790102e-03 0.9920084
#> 19 0.09621791 28 90 1.313007e-05 3.171817e-03 0.9920084
#> 20 0.09619099 12 80 1.320374e-05 3.182526e-03 0.9920084
#> 21 0.09576091 89 95 1.443542e-05 3.354777e-03 0.9891317
#> 22 0.09473210 7 51 1.784126e-05 3.864825e-03 0.9891317
#> 23 -0.09386896 53 58 2.127622e-05 4.313590e-03 0.9891317
#> 24 -0.09366615 29 83 2.217013e-05 4.421099e-03 0.9891317
#> 25 -0.09341148 21 89 2.334321e-05 4.556947e-03 0.9810727
#> 26 -0.09156391 49 93 3.380043e-05 5.955972e-03 0.9810727
#> 27 -0.09150710 80 90 3.418363e-05 6.002083e-03 0.9810727
#> 28 0.09101505 7 53 3.767966e-05 6.408102e-03 0.9810727
#> 29 0.09050688 21 84 4.164471e-05 6.838782e-03 0.9810727
#> 30 0.08965490 72 73 4.919365e-05 7.581866e-03 0.9810727
#> 31 -0.08934025 29 99 5.229604e-05 7.861416e-03 0.9810727
#> 32 -0.08906819 9 95 5.512708e-05 8.104759e-03 0.9810727
#> 33 0.08888345 2 49 5.713144e-05 8.270673e-03 0.9810727
#> 34 0.08850681 86 90 6.143363e-05 8.610161e-03 0.9810727
#> 35 0.08805868 17 53 6.695170e-05 9.015175e-03 0.9810727
#> 36 0.08790809 28 48 6.890884e-05 9.151291e-03 0.9810727
#> 37 0.08783471 33 58 6.988211e-05 9.217597e-03 0.9682377
#> 38 -0.08705796 7 49 8.101244e-05 1.021362e-02 0.9682377
#> 39 0.08645033 20 46 9.086547e-05 1.102466e-02 0.9682377
#> 40 0.08609950 48 86 9.705862e-05 1.150392e-02 0.9682377
#> 41 0.08598769 21 52 9.911458e-05 1.165816e-02 0.9682377
#> 42 0.08555275 32 95 1.075099e-04 1.226435e-02 0.9682377
#> 43 0.08548231 17 51 1.089311e-04 1.236337e-02 0.9424721
#> 44 0.08470370 80 83 1.258659e-04 1.382356e-02 0.9424721
#> 45 0.08442510 80 82 1.325062e-04 1.437068e-02 0.9174573
#> 46 0.08271606 80 93 1.810275e-04 1.845632e-02 0.9174573
#> 47 0.08235175 46 91 1.933329e-04 1.941579e-02 0.9174573
#> 48 0.08217787 25 95 1.994788e-04 1.988432e-02 0.9174573
#> 49 -0.08170331 29 87 2.171999e-04 2.119715e-02 0.9174573
#> 50 0.08123632 19 29 2.360716e-04 2.253606e-02 0.9174573
#> 51 0.08101702 51 84 2.454547e-04 2.318024e-02 0.9174573
#> 52 0.08030748 16 93 2.782643e-04 2.532796e-02 0.9174573
#> 53 0.08006503 28 52 2.903870e-04 2.608271e-02 0.9174573
#> 54 -0.07941656 41 80 3.252833e-04 2.814824e-02 0.9174573
#> 55 0.07941410 54 89 3.254229e-04 2.815620e-02 0.9174573
#> 56 -0.07934653 28 80 3.292784e-04 2.837511e-02 0.9174573
#> 57 0.07916783 29 92 3.396802e-04 2.895702e-02 0.9174573
#> 58 -0.07866905 17 86 3.703635e-04 3.060293e-02 0.9174573
#> 59 0.07827749 16 29 3.962446e-04 3.191462e-02 0.9174573
#> 60 -0.07808262 73 89 4.097452e-04 3.257290e-02 0.9174573
#> 61 0.07766261 52 67 4.403165e-04 3.400207e-02 0.9174573
#> 62 0.07762917 25 87 4.428396e-04 3.411637e-02 0.9174573
#> 63 -0.07739378 9 93 4.609872e-04 3.492295e-02 0.9174573
#> 64 0.07738885 31 80 4.613747e-04 3.493988e-02 0.9174573
#> 65 -0.07718681 80 94 4.775136e-04 3.563444e-02 0.9174573
#> 66 0.07706275 27 58 4.876831e-04 3.606179e-02 0.8297811
#> 67 -0.07610709 16 83 5.730532e-04 4.085920e-02 0.8297811
#> 68 0.07550557 53 84 6.337143e-04 4.406472e-02 0.8297811
# significant based on "local fdr" cutoff (prob > 0.9)?
num.significant.2 <- sum(test.results$prob > 0.9)
test.results[test.results$prob > 0.9,]
#> pcor node1 node2 pval qval prob
#> 1 0.23185664 51 53 2.220446e-16 3.612205e-13 1.0000000
#> 2 0.22405545 52 53 2.220446e-16 3.612205e-13 1.0000000
#> 3 0.21507824 51 52 2.220446e-16 3.612205e-13 1.0000000
#> 4 0.17328863 7 93 3.108624e-15 3.792816e-12 0.9999945
#> 5 -0.13418892 29 86 1.120812e-09 1.093997e-06 0.9999516
#> 6 0.12594697 21 72 1.103836e-08 8.978563e-06 0.9998400
#> 7 0.11956105 28 86 5.890924e-08 3.853590e-05 0.9998400
#> 8 -0.11723897 26 80 1.060526e-07 5.816172e-05 0.9998400
#> 9 -0.11711625 72 89 1.093655e-07 5.930499e-05 0.9972804
#> 10 0.10658013 20 21 1.366610e-06 5.925275e-04 0.9972804
#> 11 0.10589778 21 73 1.596859e-06 6.678429e-04 0.9972804
#> 12 0.10478689 20 91 2.053403e-06 8.024425e-04 0.9972804
#> 13 0.10420836 7 52 2.338382e-06 8.778605e-04 0.9944557
#> 14 0.10236077 87 95 3.525186e-06 1.224964e-03 0.9944557
#> 15 0.10113550 27 95 4.610444e-06 1.500047e-03 0.9920084
#> 16 0.09928954 21 51 6.868357e-06 2.046549e-03 0.9920084
#> 17 0.09791914 21 88 9.192373e-06 2.520616e-03 0.9920084
#> 18 0.09719685 18 95 1.070232e-05 2.790102e-03 0.9920084
#> 19 0.09621791 28 90 1.313007e-05 3.171817e-03 0.9920084
#> 20 0.09619099 12 80 1.320374e-05 3.182526e-03 0.9920084
#> 21 0.09576091 89 95 1.443542e-05 3.354777e-03 0.9891317
#> 22 0.09473210 7 51 1.784126e-05 3.864825e-03 0.9891317
#> 23 -0.09386896 53 58 2.127622e-05 4.313590e-03 0.9891317
#> 24 -0.09366615 29 83 2.217013e-05 4.421099e-03 0.9891317
#> 25 -0.09341148 21 89 2.334321e-05 4.556947e-03 0.9810727
#> 26 -0.09156391 49 93 3.380043e-05 5.955972e-03 0.9810727
#> 27 -0.09150710 80 90 3.418363e-05 6.002083e-03 0.9810727
#> 28 0.09101505 7 53 3.767966e-05 6.408102e-03 0.9810727
#> 29 0.09050688 21 84 4.164471e-05 6.838782e-03 0.9810727
#> 30 0.08965490 72 73 4.919365e-05 7.581866e-03 0.9810727
#> 31 -0.08934025 29 99 5.229604e-05 7.861416e-03 0.9810727
#> 32 -0.08906819 9 95 5.512708e-05 8.104759e-03 0.9810727
#> 33 0.08888345 2 49 5.713144e-05 8.270673e-03 0.9810727
#> 34 0.08850681 86 90 6.143363e-05 8.610161e-03 0.9810727
#> 35 0.08805868 17 53 6.695170e-05 9.015175e-03 0.9810727
#> 36 0.08790809 28 48 6.890884e-05 9.151291e-03 0.9810727
#> 37 0.08783471 33 58 6.988211e-05 9.217597e-03 0.9682377
#> 38 -0.08705796 7 49 8.101244e-05 1.021362e-02 0.9682377
#> 39 0.08645033 20 46 9.086547e-05 1.102466e-02 0.9682377
#> 40 0.08609950 48 86 9.705862e-05 1.150392e-02 0.9682377
#> 41 0.08598769 21 52 9.911458e-05 1.165816e-02 0.9682377
#> 42 0.08555275 32 95 1.075099e-04 1.226435e-02 0.9682377
#> 43 0.08548231 17 51 1.089311e-04 1.236337e-02 0.9424721
#> 44 0.08470370 80 83 1.258659e-04 1.382356e-02 0.9424721
#> 45 0.08442510 80 82 1.325062e-04 1.437068e-02 0.9174573
#> 46 0.08271606 80 93 1.810275e-04 1.845632e-02 0.9174573
#> 47 0.08235175 46 91 1.933329e-04 1.941579e-02 0.9174573
#> 48 0.08217787 25 95 1.994788e-04 1.988432e-02 0.9174573
#> 49 -0.08170331 29 87 2.171999e-04 2.119715e-02 0.9174573
#> 50 0.08123632 19 29 2.360716e-04 2.253606e-02 0.9174573
#> 51 0.08101702 51 84 2.454547e-04 2.318024e-02 0.9174573
#> 52 0.08030748 16 93 2.782643e-04 2.532796e-02 0.9174573
#> 53 0.08006503 28 52 2.903870e-04 2.608271e-02 0.9174573
#> 54 -0.07941656 41 80 3.252833e-04 2.814824e-02 0.9174573
#> 55 0.07941410 54 89 3.254229e-04 2.815620e-02 0.9174573
#> 56 -0.07934653 28 80 3.292784e-04 2.837511e-02 0.9174573
#> 57 0.07916783 29 92 3.396802e-04 2.895702e-02 0.9174573
#> 58 -0.07866905 17 86 3.703635e-04 3.060293e-02 0.9174573
#> 59 0.07827749 16 29 3.962446e-04 3.191462e-02 0.9174573
#> 60 -0.07808262 73 89 4.097452e-04 3.257290e-02 0.9174573
#> 61 0.07766261 52 67 4.403165e-04 3.400207e-02 0.9174573
#> 62 0.07762917 25 87 4.428396e-04 3.411637e-02 0.9174573
#> 63 -0.07739378 9 93 4.609872e-04 3.492295e-02 0.9174573
#> 64 0.07738885 31 80 4.613747e-04 3.493988e-02 0.9174573
#> 65 -0.07718681 80 94 4.775136e-04 3.563444e-02 0.9174573
# parameters of the mixture distribution used to compute p-values etc.
c <- fdrtool::fdrtool(corpcor::sm2vec(inferred.pcor), statistic="correlation")
#> Step 1... determine cutoff point
#> Step 2... estimate parameters of null distribution and eta0
#> Step 3... compute p-values and estimate empirical PDF/CDF
#> Step 4... compute q-values and local fdr
#> Step 5... prepare for plotting
c$param
#> cutoff N.cens eta0 eta0.SE kappa kappa.SE
#> [1,] 0.03553068 4352 0.9474623 0.005656465 2043.377 94.72267
## A random network with 20 nodes and 10 percent (=19) edges
true.pcor <- GeneNet::ggm.simulate.pcor(20, 0.1)
# convert to edge list
test.results <- GeneNet::ggm.list.edges(true.pcor)[1:19,]
nlab <- LETTERS[1:20]
# graphviz
# network.make.dot(filename="test.dot", test.results, nlab, main = "A graph")
# system("fdp -T svg -o test.svg test.dot")
# Rgraphviz
gr <- GeneNet::network.make.graph( test.results, nlab)
gr
#> A graphNEL graph with directed edges
#> Number of Nodes = 20
#> Number of Edges = 38
num.nodes(gr)
#> [1] 20
edge.info(gr)
#> $weight
#> A~H D~F D~O E~R F~L F~G G~R G~O
#> -0.76325 -0.47042 -0.53865 -0.52631 -0.05494 0.42383 -0.19385 0.32270
#> H~T I~J J~S J~M K~R K~L L~N N~R
#> -0.34139 0.54040 0.41751 0.62051 0.33471 -0.64564 -0.05633 0.55074
#> O~R O~P S~T
#> -0.04365 -0.37113 0.57545
#>
#> $dir
#> A~H D~F D~O E~R F~L F~G G~R G~O H~T I~J J~S
#> "none" "none" "none" "none" "none" "none" "none" "none" "none" "none" "none"
#> J~M K~R K~L L~N N~R O~R O~P S~T
#> "none" "none" "none" "none" "none" "none" "none" "none"
gr2 <- GeneNet::network.make.graph( test.results, nlab, drop.singles=TRUE)
gr2
#> A graphNEL graph with directed edges
#> Number of Nodes = 17
#> Number of Edges = 38
GeneNet::num.nodes(gr2)
#> [1] 17
GeneNet::edge.info(gr2)
#> $weight
#> A~H D~F D~O E~R F~L F~G G~R G~O
#> -0.76325 -0.47042 -0.53865 -0.52631 -0.05494 0.42383 -0.19385 0.32270
#> H~T I~J J~S J~M K~R K~L L~N N~R
#> -0.34139 0.54040 0.41751 0.62051 0.33471 -0.64564 -0.05633 0.55074
#> O~R O~P S~T
#> -0.04365 -0.37113 0.57545
#>
#> $dir
#> A~H D~F D~O E~R F~L F~G G~R G~O H~T I~J J~S
#> "none" "none" "none" "none" "none" "none" "none" "none" "none" "none" "none"
#> J~M K~R K~L L~N N~R O~R O~P S~T
#> "none" "none" "none" "none" "none" "none" "none" "none"
# plot network
plot(gr, "fdp")
#> Warning in arrows(tail_from[1], tail_from[2], tail_to[1], tail_to[2], col =
#> edgeColor, : zero-length arrow is of indeterminate angle and so skipped
#> Warning in arrows(head_from[1], head_from[2], head_to[1], head_to[2], col =
#> edgeColor, : zero-length arrow is of indeterminate angle and so skipped
#> Warning in arrows(head_from[1], head_from[2], head_to[1], head_to[2], col =
#> edgeColor, : zero-length arrow is of indeterminate angle and so skipped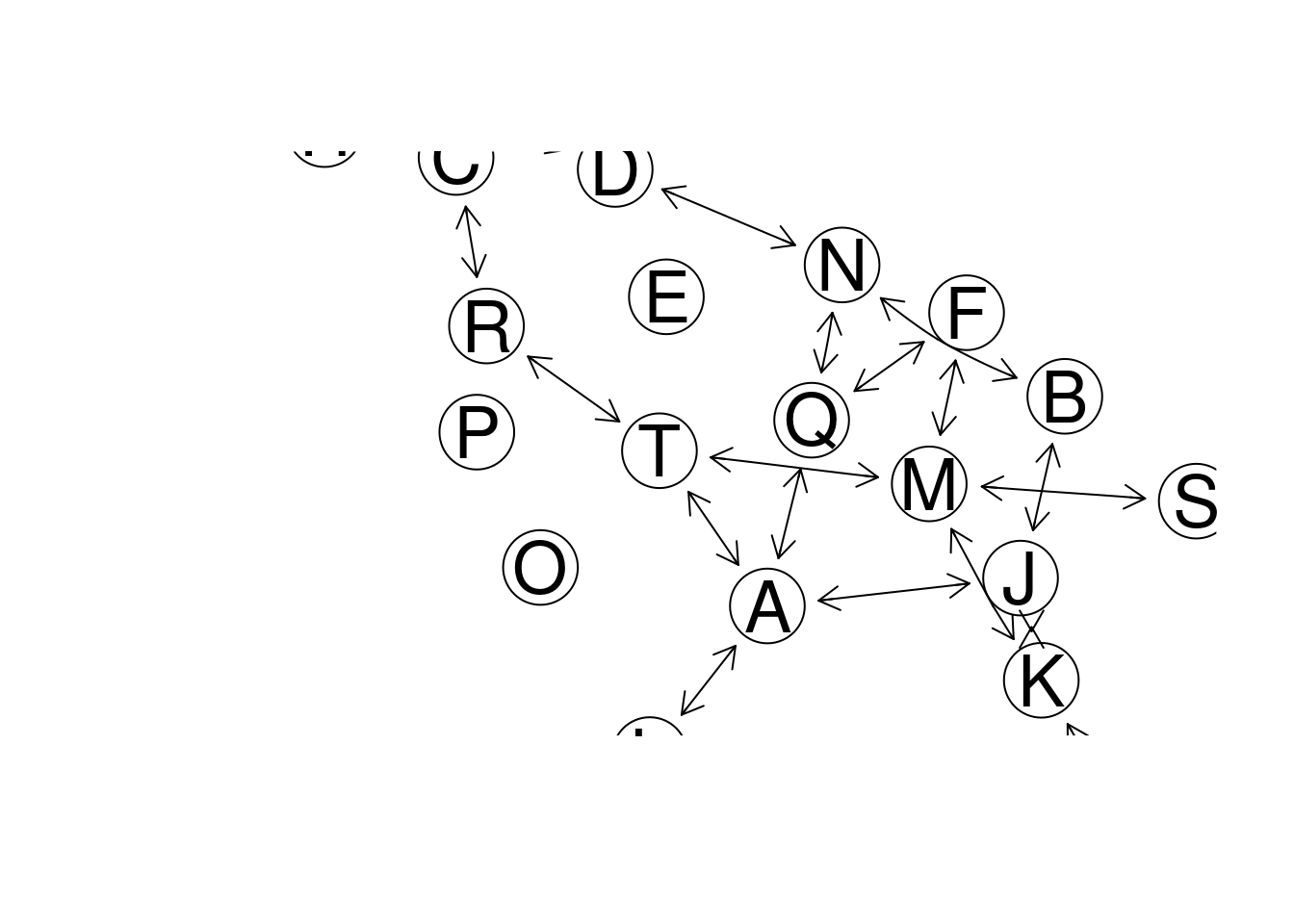
Figure 7.2: GeneNet example
plot(gr2, "fdp")
#> Warning in arrows(tail_from[1], tail_from[2], tail_to[1], tail_to[2], col =
#> edgeColor, : zero-length arrow is of indeterminate angle and so skipped
#> Warning in arrows(tail_from[1], tail_from[2], tail_to[1], tail_to[2], col =
#> edgeColor, : zero-length arrow is of indeterminate angle and so skipped
#> Warning in arrows(tail_from[1], tail_from[2], tail_to[1], tail_to[2], col =
#> edgeColor, : zero-length arrow is of indeterminate angle and so skipped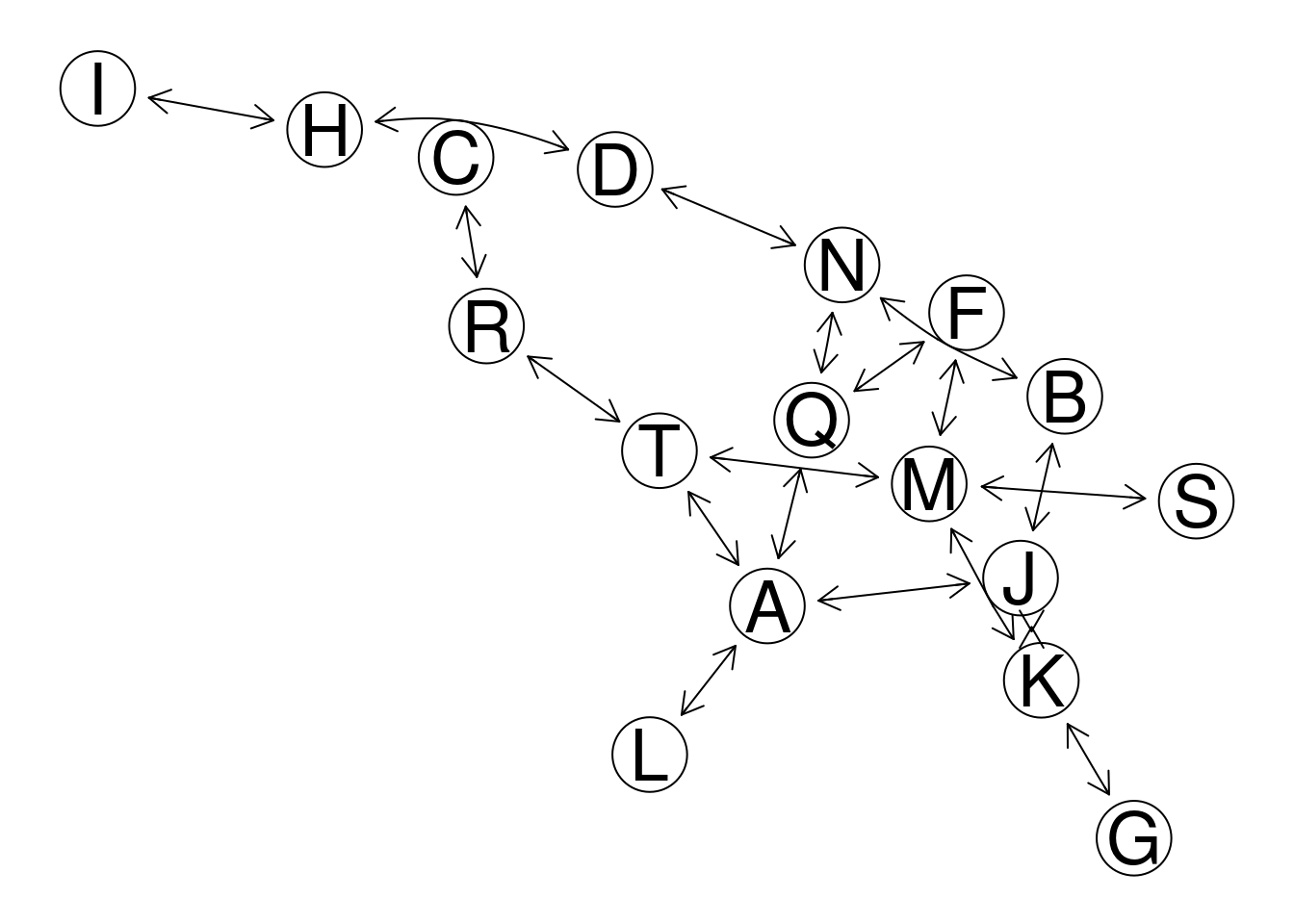
Figure 7.3: GeneNet example
A side-by-side heatmaps
set.seed(123454321)
m <- matrix(runif(2500),50)
r <- cor(m)
g <- as.matrix(r>=0.7)+0
f1 <- ComplexHeatmap::Heatmap(r)
f2 <- ComplexHeatmap::Heatmap(g)
f <- f1+f2
ComplexHeatmap::draw(f)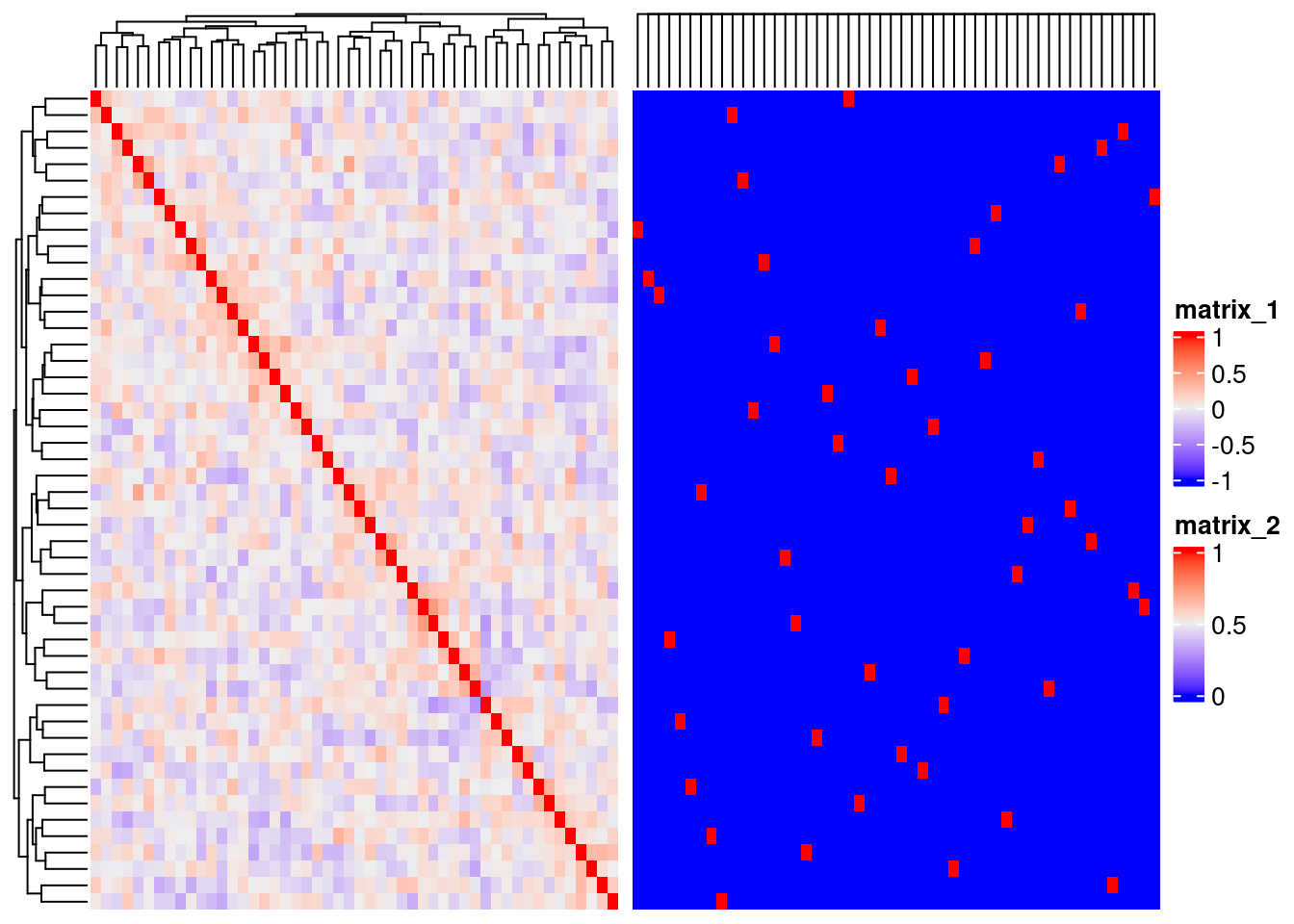
Figure 7.4: Heatmaps
df <- heatmaply::normalize(mtcars)
hm <- heatmaply::heatmaply(df,k_col=5,k_row=5,
colors = grDevices::colorRampPalette(RColorBrewer::brewer.pal(3, "RdBu"))(256))
htmlwidgets::saveWidget(hm,file="heatmaply.html")
htmltools::tags$iframe(src = "heatmaply.html", width = "100%", height = "550px")so we have heatmaply.html and a module analysis with WGCNA,
pwr <- c(1:10, seq(from=12, to=30, by=2))
sft <- WGCNA::pickSoftThreshold(dat, powerVector=pwr, verbose=5)
ADJ <- abs(cor(dat, method="pearson", use="pairwise.complete.obs"))^6
dissADJ <- 1-ADJ
dissTOM <- WGCNA::TOMdist(ADJ)
TOM <- WGCNA::TOMsimilarityFromExpr(dat)
Tree <- hclust(as.dist(1-TOM), method="average")
for(j in pwr)
{
pam_name <- paste0("pam",j)
assign(pam_name, cluster::pam(as.dist(dissADJ),j))
pamTOM_name <- paste0("pamTOM",j)
assign(pamTOM_name,cluster::pam(as.dist(dissTOM),j))
tc <- table(get(pam_name)$clustering,get(pamTOM_name)$clustering)
print(tc)
print(diag(tc))
}
colorStaticTOM <- as.character(WGCNA::cutreeStaticColor(Tree,cutHeight=.99,minSize=5))
colorDynamicTOM <- WGCNA::labels2colors(cutreeDynamic(Tree,method="tree",minClusterSize=5))
Colors <- data.frame(pamTOM6$clustering,colorStaticTOM,colorDynamicTOM)
WGCNA::plotDendroAndColors(Tree, Colors, dendroLabels=FALSE, hang=0.03, addGuide=TRUE, guideHang=0.05)
meg <- WGCNA::moduleEigengenes(dat, color=1:ncol(dat), softPower=6)8 Metadata
This section is based on package recount3.
hs <- recount3::available_projects()
dim(subset(hs,file_source=="gtex"))
recount3::annotation_options("human")
blood_rse <- recount3::create_rse(subset(hs,project=="BLOOD"))
S4Vectors::metadata(blood_rse)
SummarizedExperiment::rowRanges(blood_rse)
colnames(SummarizedExperiment::colData(blood_rse))[1:20]
recount3::expand_sra_attributes(blood_rse)9 Pathway and enrichment analysis
reactome <- graphite::pathways("hsapiens", "reactome")
kegg <- graphite::pathways("hsapiens","kegg")
pharmgkb <- graphite::pathways("hsapiens","pharmgkb")
nodes(kegg[[21]])
#> [1] "ENTREZID:102724560" "ENTREZID:10993" "ENTREZID:113675"
#> [4] "ENTREZID:132158" "ENTREZID:1610" "ENTREZID:1738"
#> [7] "ENTREZID:1757" "ENTREZID:189" "ENTREZID:211"
#> [10] "ENTREZID:212" "ENTREZID:23464" "ENTREZID:2593"
#> [13] "ENTREZID:26227" "ENTREZID:2628" "ENTREZID:27232"
#> [16] "ENTREZID:2731" "ENTREZID:275" "ENTREZID:29958"
#> [19] "ENTREZID:29968" "ENTREZID:441531" "ENTREZID:501"
#> [22] "ENTREZID:51268" "ENTREZID:5223" "ENTREZID:5224"
#> [25] "ENTREZID:55349" "ENTREZID:5723" "ENTREZID:635"
#> [28] "ENTREZID:63826" "ENTREZID:6470" "ENTREZID:6472"
#> [31] "ENTREZID:64902" "ENTREZID:669" "ENTREZID:875"
#> [34] "ENTREZID:9380" "ENTREZID:1491"
kegg_t2g <- ldply(lapply(kegg, nodes), data.frame)
names(kegg_t2g) <- c("gs_name", "gene_symbol")
VEGF <- subset(kegg_t2g,gs_name=="VEGF signaling pathway")[[2]]
eKEGG <- clusterProfiler::enricher(gene=VEGF, TERM2GENE = kegg_t2g,
universe=,
pAdjustMethod = "BH",
pvalueCutoff = 0.1, qvalueCutoff = 0.05,
minGSSize = 10, maxGSSize = 500)10 Transcript databases
An overview of annotation is available2.
# columns(org.Hs.eg.db)
# keyref <- keys(org.Hs.eg.db, keytype="ENTREZID")
# symbol_uniprot <- select(org.Hs.eg.db,keys=keyref,columns = c("SYMBOL","UNIPROT"))
# subset(symbol_uniprot,SYMBOL=="MC4R")
x <- EnsDb.Hsapiens.v86
ensembldb::listColumns(x, "protein", skip.keys=TRUE)
#> [1] "tx_id" "protein_id" "protein_sequence"
ensembldb::listGenebiotypes(x)
#> [1] "protein_coding" "unitary_pseudogene"
#> [3] "unprocessed_pseudogene" "processed_pseudogene"
#> [5] "processed_transcript" "transcribed_unprocessed_pseudogene"
#> [7] "antisense" "transcribed_unitary_pseudogene"
#> [9] "polymorphic_pseudogene" "lincRNA"
#> [11] "sense_intronic" "transcribed_processed_pseudogene"
#> [13] "sense_overlapping" "IG_V_pseudogene"
#> [15] "pseudogene" "TR_V_gene"
#> [17] "3prime_overlapping_ncRNA" "IG_V_gene"
#> [19] "bidirectional_promoter_lncRNA" "snRNA"
#> [21] "miRNA" "misc_RNA"
#> [23] "snoRNA" "rRNA"
#> [25] "Mt_tRNA" "Mt_rRNA"
#> [27] "IG_C_gene" "IG_J_gene"
#> [29] "TR_J_gene" "TR_C_gene"
#> [31] "TR_V_pseudogene" "TR_J_pseudogene"
#> [33] "IG_D_gene" "ribozyme"
#> [35] "IG_C_pseudogene" "TR_D_gene"
#> [37] "TEC" "IG_J_pseudogene"
#> [39] "scRNA" "scaRNA"
#> [41] "vaultRNA" "sRNA"
#> [43] "macro_lncRNA" "non_coding"
#> [45] "IG_pseudogene" "LRG_gene"
ensembldb::listTxbiotypes(x)
#> [1] "protein_coding" "processed_transcript"
#> [3] "nonsense_mediated_decay" "retained_intron"
#> [5] "unitary_pseudogene" "TEC"
#> [7] "miRNA" "misc_RNA"
#> [9] "non_stop_decay" "unprocessed_pseudogene"
#> [11] "processed_pseudogene" "transcribed_unprocessed_pseudogene"
#> [13] "lincRNA" "antisense"
#> [15] "transcribed_unitary_pseudogene" "polymorphic_pseudogene"
#> [17] "sense_intronic" "transcribed_processed_pseudogene"
#> [19] "sense_overlapping" "IG_V_pseudogene"
#> [21] "pseudogene" "TR_V_gene"
#> [23] "3prime_overlapping_ncRNA" "IG_V_gene"
#> [25] "bidirectional_promoter_lncRNA" "snRNA"
#> [27] "snoRNA" "rRNA"
#> [29] "Mt_tRNA" "Mt_rRNA"
#> [31] "IG_C_gene" "IG_J_gene"
#> [33] "TR_J_gene" "TR_C_gene"
#> [35] "TR_V_pseudogene" "TR_J_pseudogene"
#> [37] "IG_D_gene" "ribozyme"
#> [39] "IG_C_pseudogene" "TR_D_gene"
#> [41] "IG_J_pseudogene" "scRNA"
#> [43] "scaRNA" "vaultRNA"
#> [45] "sRNA" "macro_lncRNA"
#> [47] "non_coding" "IG_pseudogene"
#> [49] "LRG_gene"
ensembldb::listTables(x)
#> $gene
#> [1] "gene_id" "gene_name" "gene_biotype" "gene_seq_start"
#> [5] "gene_seq_end" "seq_name" "seq_strand" "seq_coord_system"
#> [9] "symbol"
#>
#> $tx
#> [1] "tx_id" "tx_biotype" "tx_seq_start" "tx_seq_end"
#> [5] "tx_cds_seq_start" "tx_cds_seq_end" "gene_id" "tx_name"
#>
#> $tx2exon
#> [1] "tx_id" "exon_id" "exon_idx"
#>
#> $exon
#> [1] "exon_id" "exon_seq_start" "exon_seq_end"
#>
#> $chromosome
#> [1] "seq_name" "seq_length" "is_circular"
#>
#> $protein
#> [1] "tx_id" "protein_id" "protein_sequence"
#>
#> $uniprot
#> [1] "protein_id" "uniprot_id" "uniprot_db"
#> [4] "uniprot_mapping_type"
#>
#> $protein_domain
#> [1] "protein_id" "protein_domain_id" "protein_domain_source"
#> [4] "interpro_accession" "prot_dom_start" "prot_dom_end"
#>
#> $entrezgene
#> [1] "gene_id" "entrezid"
#>
#> $metadata
#> [1] "name" "value"
ensembldb::metadata(x)
#> name value
#> 1 Db type EnsDb
#> 2 Type of Gene ID Ensembl Gene ID
#> 3 Supporting package ensembldb
#> 4 Db created by ensembldb package from Bioconductor
#> 5 script_version 0.3.0
#> 6 Creation time Thu May 18 16:32:27 2017
#> 7 ensembl_version 86
#> 8 ensembl_host localhost
#> 9 Organism homo_sapiens
#> 10 taxonomy_id 9606
#> 11 genome_build GRCh38
#> 12 DBSCHEMAVERSION 2.0
ensembldb::organism(x)
#> [1] "Homo sapiens"
ensembldb::returnFilterColumns(x)
#> [1] TRUE
ensembldb::seqinfo(x)
#> Seqinfo object with 357 sequences (1 circular) from GRCh38 genome:
#> seqnames seqlengths isCircular genome
#> X 156040895 FALSE GRCh38
#> 20 64444167 FALSE GRCh38
#> 1 248956422 FALSE GRCh38
#> 6 170805979 FALSE GRCh38
#> 3 198295559 FALSE GRCh38
#> ... ... ... ...
#> LRG_239 114904 FALSE GRCh38
#> LRG_311 115492 FALSE GRCh38
#> LRG_721 33396 FALSE GRCh38
#> LRG_741 231167 FALSE GRCh38
#> LRG_93 22459 FALSE GRCh38
ensembldb::seqlevels(x)
#> [1] "1"
#> [2] "10"
#> [3] "11"
#> [4] "12"
#> [5] "13"
#> [6] "14"
#> [7] "15"
#> [8] "16"
#> [9] "17"
#> [10] "18"
#> [11] "19"
#> [12] "2"
#> [13] "20"
#> [14] "21"
#> [15] "22"
#> [16] "3"
#> [17] "4"
#> [18] "5"
#> [19] "6"
#> [20] "7"
#> [21] "8"
#> [22] "9"
#> [23] "CHR_HG107_PATCH"
#> [24] "CHR_HG126_PATCH"
#> [25] "CHR_HG1311_PATCH"
#> [26] "CHR_HG1342_HG2282_PATCH"
#> [27] "CHR_HG1362_PATCH"
#> [28] "CHR_HG142_HG150_NOVEL_TEST"
#> [29] "CHR_HG151_NOVEL_TEST"
#> [30] "CHR_HG1651_PATCH"
#> [31] "CHR_HG1832_PATCH"
#> [32] "CHR_HG2021_PATCH"
#> [33] "CHR_HG2022_PATCH"
#> [34] "CHR_HG2023_PATCH"
#> [35] "CHR_HG2030_PATCH"
#> [36] "CHR_HG2058_PATCH"
#> [37] "CHR_HG2062_PATCH"
#> [38] "CHR_HG2063_PATCH"
#> [39] "CHR_HG2066_PATCH"
#> [40] "CHR_HG2072_PATCH"
#> [41] "CHR_HG2095_PATCH"
#> [42] "CHR_HG2104_PATCH"
#> [43] "CHR_HG2116_PATCH"
#> [44] "CHR_HG2128_PATCH"
#> [45] "CHR_HG2191_PATCH"
#> [46] "CHR_HG2213_PATCH"
#> [47] "CHR_HG2217_PATCH"
#> [48] "CHR_HG2232_PATCH"
#> [49] "CHR_HG2233_PATCH"
#> [50] "CHR_HG2235_PATCH"
#> [51] "CHR_HG2239_PATCH"
#> [52] "CHR_HG2247_PATCH"
#> [53] "CHR_HG2249_PATCH"
#> [54] "CHR_HG2288_HG2289_PATCH"
#> [55] "CHR_HG2290_PATCH"
#> [56] "CHR_HG2291_PATCH"
#> [57] "CHR_HG2334_PATCH"
#> [58] "CHR_HG26_PATCH"
#> [59] "CHR_HG986_PATCH"
#> [60] "CHR_HSCHR10_1_CTG1"
#> [61] "CHR_HSCHR10_1_CTG2"
#> [62] "CHR_HSCHR10_1_CTG3"
#> [63] "CHR_HSCHR10_1_CTG4"
#> [64] "CHR_HSCHR10_1_CTG6"
#> [65] "CHR_HSCHR11_1_CTG1_2"
#> [66] "CHR_HSCHR11_1_CTG5"
#> [67] "CHR_HSCHR11_1_CTG6"
#> [68] "CHR_HSCHR11_1_CTG7"
#> [69] "CHR_HSCHR11_1_CTG8"
#> [70] "CHR_HSCHR11_2_CTG1"
#> [71] "CHR_HSCHR11_2_CTG1_1"
#> [72] "CHR_HSCHR11_3_CTG1"
#> [73] "CHR_HSCHR12_1_CTG1"
#> [74] "CHR_HSCHR12_1_CTG2_1"
#> [75] "CHR_HSCHR12_2_CTG1"
#> [76] "CHR_HSCHR12_2_CTG2"
#> [77] "CHR_HSCHR12_2_CTG2_1"
#> [78] "CHR_HSCHR12_3_CTG2"
#> [79] "CHR_HSCHR12_3_CTG2_1"
#> [80] "CHR_HSCHR12_4_CTG2"
#> [81] "CHR_HSCHR12_4_CTG2_1"
#> [82] "CHR_HSCHR12_5_CTG2"
#> [83] "CHR_HSCHR12_5_CTG2_1"
#> [84] "CHR_HSCHR12_6_CTG2_1"
#> [85] "CHR_HSCHR13_1_CTG1"
#> [86] "CHR_HSCHR13_1_CTG3"
#> [87] "CHR_HSCHR13_1_CTG5"
#> [88] "CHR_HSCHR13_1_CTG8"
#> [89] "CHR_HSCHR14_1_CTG1"
#> [90] "CHR_HSCHR14_2_CTG1"
#> [91] "CHR_HSCHR14_3_CTG1"
#> [92] "CHR_HSCHR14_7_CTG1"
#> [93] "CHR_HSCHR15_1_CTG1"
#> [94] "CHR_HSCHR15_1_CTG3"
#> [95] "CHR_HSCHR15_1_CTG8"
#> [96] "CHR_HSCHR15_2_CTG3"
#> [97] "CHR_HSCHR15_2_CTG8"
#> [98] "CHR_HSCHR15_3_CTG3"
#> [99] "CHR_HSCHR15_3_CTG8"
#> [100] "CHR_HSCHR15_4_CTG8"
#> [101] "CHR_HSCHR15_5_CTG8"
#> [102] "CHR_HSCHR15_6_CTG8"
#> [103] "CHR_HSCHR16_1_CTG1"
#> [104] "CHR_HSCHR16_1_CTG3_1"
#> [105] "CHR_HSCHR16_2_CTG3_1"
#> [106] "CHR_HSCHR16_3_CTG1"
#> [107] "CHR_HSCHR16_4_CTG1"
#> [108] "CHR_HSCHR16_4_CTG3_1"
#> [109] "CHR_HSCHR16_5_CTG1"
#> [110] "CHR_HSCHR16_CTG2"
#> [111] "CHR_HSCHR17_10_CTG4"
#> [112] "CHR_HSCHR17_1_CTG1"
#> [113] "CHR_HSCHR17_1_CTG2"
#> [114] "CHR_HSCHR17_1_CTG4"
#> [115] "CHR_HSCHR17_1_CTG5"
#> [116] "CHR_HSCHR17_1_CTG9"
#> [117] "CHR_HSCHR17_2_CTG1"
#> [118] "CHR_HSCHR17_2_CTG2"
#> [119] "CHR_HSCHR17_2_CTG4"
#> [120] "CHR_HSCHR17_2_CTG5"
#> [121] "CHR_HSCHR17_3_CTG2"
#> [122] "CHR_HSCHR17_3_CTG4"
#> [123] "CHR_HSCHR17_4_CTG4"
#> [124] "CHR_HSCHR17_5_CTG4"
#> [125] "CHR_HSCHR17_6_CTG4"
#> [126] "CHR_HSCHR17_7_CTG4"
#> [127] "CHR_HSCHR17_8_CTG4"
#> [128] "CHR_HSCHR17_9_CTG4"
#> [129] "CHR_HSCHR18_1_CTG1_1"
#> [130] "CHR_HSCHR18_1_CTG2_1"
#> [131] "CHR_HSCHR18_2_CTG1_1"
#> [132] "CHR_HSCHR18_2_CTG2"
#> [133] "CHR_HSCHR18_2_CTG2_1"
#> [134] "CHR_HSCHR18_3_CTG2_1"
#> [135] "CHR_HSCHR18_5_CTG1_1"
#> [136] "CHR_HSCHR18_ALT21_CTG2_1"
#> [137] "CHR_HSCHR18_ALT2_CTG2_1"
#> [138] "CHR_HSCHR19KIR_ABC08_A1_HAP_CTG3_1"
#> [139] "CHR_HSCHR19KIR_ABC08_AB_HAP_C_P_CTG3_1"
#> [140] "CHR_HSCHR19KIR_ABC08_AB_HAP_T_P_CTG3_1"
#> [141] "CHR_HSCHR19KIR_FH05_A_HAP_CTG3_1"
#> [142] "CHR_HSCHR19KIR_FH05_B_HAP_CTG3_1"
#> [143] "CHR_HSCHR19KIR_FH06_A_HAP_CTG3_1"
#> [144] "CHR_HSCHR19KIR_FH06_BA1_HAP_CTG3_1"
#> [145] "CHR_HSCHR19KIR_FH08_A_HAP_CTG3_1"
#> [146] "CHR_HSCHR19KIR_FH08_BAX_HAP_CTG3_1"
#> [147] "CHR_HSCHR19KIR_FH13_A_HAP_CTG3_1"
#> [148] "CHR_HSCHR19KIR_FH13_BA2_HAP_CTG3_1"
#> [149] "CHR_HSCHR19KIR_FH15_A_HAP_CTG3_1"
#> [150] "CHR_HSCHR19KIR_FH15_B_HAP_CTG3_1"
#> [151] "CHR_HSCHR19KIR_G085_A_HAP_CTG3_1"
#> [152] "CHR_HSCHR19KIR_G085_BA1_HAP_CTG3_1"
#> [153] "CHR_HSCHR19KIR_G248_A_HAP_CTG3_1"
#> [154] "CHR_HSCHR19KIR_G248_BA2_HAP_CTG3_1"
#> [155] "CHR_HSCHR19KIR_GRC212_AB_HAP_CTG3_1"
#> [156] "CHR_HSCHR19KIR_GRC212_BA1_HAP_CTG3_1"
#> [157] "CHR_HSCHR19KIR_LUCE_A_HAP_CTG3_1"
#> [158] "CHR_HSCHR19KIR_LUCE_BDEL_HAP_CTG3_1"
#> [159] "CHR_HSCHR19KIR_RP5_B_HAP_CTG3_1"
#> [160] "CHR_HSCHR19KIR_RSH_A_HAP_CTG3_1"
#> [161] "CHR_HSCHR19KIR_RSH_BA2_HAP_CTG3_1"
#> [162] "CHR_HSCHR19KIR_T7526_A_HAP_CTG3_1"
#> [163] "CHR_HSCHR19KIR_T7526_BDEL_HAP_CTG3_1"
#> [164] "CHR_HSCHR19LRC_COX1_CTG3_1"
#> [165] "CHR_HSCHR19LRC_COX2_CTG3_1"
#> [166] "CHR_HSCHR19LRC_LRC_I_CTG3_1"
#> [167] "CHR_HSCHR19LRC_LRC_J_CTG3_1"
#> [168] "CHR_HSCHR19LRC_LRC_S_CTG3_1"
#> [169] "CHR_HSCHR19LRC_LRC_T_CTG3_1"
#> [170] "CHR_HSCHR19LRC_PGF1_CTG3_1"
#> [171] "CHR_HSCHR19LRC_PGF2_CTG3_1"
#> [172] "CHR_HSCHR19_1_CTG2"
#> [173] "CHR_HSCHR19_1_CTG3_1"
#> [174] "CHR_HSCHR19_2_CTG2"
#> [175] "CHR_HSCHR19_2_CTG3_1"
#> [176] "CHR_HSCHR19_3_CTG2"
#> [177] "CHR_HSCHR19_3_CTG3_1"
#> [178] "CHR_HSCHR19_4_CTG2"
#> [179] "CHR_HSCHR19_4_CTG3_1"
#> [180] "CHR_HSCHR19_5_CTG2"
#> [181] "CHR_HSCHR1_1_CTG11"
#> [182] "CHR_HSCHR1_1_CTG3"
#> [183] "CHR_HSCHR1_1_CTG31"
#> [184] "CHR_HSCHR1_1_CTG32_1"
#> [185] "CHR_HSCHR1_2_CTG3"
#> [186] "CHR_HSCHR1_2_CTG31"
#> [187] "CHR_HSCHR1_2_CTG32_1"
#> [188] "CHR_HSCHR1_3_CTG3"
#> [189] "CHR_HSCHR1_3_CTG31"
#> [190] "CHR_HSCHR1_3_CTG32_1"
#> [191] "CHR_HSCHR1_4_CTG3"
#> [192] "CHR_HSCHR1_4_CTG31"
#> [193] "CHR_HSCHR1_5_CTG3"
#> [194] "CHR_HSCHR1_5_CTG32_1"
#> [195] "CHR_HSCHR1_ALT2_1_CTG32_1"
#> [196] "CHR_HSCHR20_1_CTG1"
#> [197] "CHR_HSCHR20_1_CTG2"
#> [198] "CHR_HSCHR20_1_CTG3"
#> [199] "CHR_HSCHR20_1_CTG4"
#> [200] "CHR_HSCHR21_2_CTG1_1"
#> [201] "CHR_HSCHR21_3_CTG1_1"
#> [202] "CHR_HSCHR21_4_CTG1_1"
#> [203] "CHR_HSCHR21_5_CTG2"
#> [204] "CHR_HSCHR21_6_CTG1_1"
#> [205] "CHR_HSCHR21_8_CTG1_1"
#> [206] "CHR_HSCHR22_1_CTG1"
#> [207] "CHR_HSCHR22_1_CTG2"
#> [208] "CHR_HSCHR22_1_CTG3"
#> [209] "CHR_HSCHR22_1_CTG4"
#> [210] "CHR_HSCHR22_1_CTG5"
#> [211] "CHR_HSCHR22_1_CTG6"
#> [212] "CHR_HSCHR22_1_CTG7"
#> [213] "CHR_HSCHR22_2_CTG1"
#> [214] "CHR_HSCHR22_3_CTG1"
#> [215] "CHR_HSCHR22_4_CTG1"
#> [216] "CHR_HSCHR22_5_CTG1"
#> [217] "CHR_HSCHR22_6_CTG1"
#> [218] "CHR_HSCHR22_7_CTG1"
#> [219] "CHR_HSCHR22_8_CTG1"
#> [220] "CHR_HSCHR2_1_CTG1"
#> [221] "CHR_HSCHR2_1_CTG15"
#> [222] "CHR_HSCHR2_1_CTG5"
#> [223] "CHR_HSCHR2_1_CTG7"
#> [224] "CHR_HSCHR2_1_CTG7_2"
#> [225] "CHR_HSCHR2_2_CTG1"
#> [226] "CHR_HSCHR2_2_CTG15"
#> [227] "CHR_HSCHR2_2_CTG7"
#> [228] "CHR_HSCHR2_2_CTG7_2"
#> [229] "CHR_HSCHR2_3_CTG1"
#> [230] "CHR_HSCHR2_3_CTG15"
#> [231] "CHR_HSCHR2_3_CTG7_2"
#> [232] "CHR_HSCHR2_4_CTG1"
#> [233] "CHR_HSCHR2_6_CTG7_2"
#> [234] "CHR_HSCHR3_1_CTG1"
#> [235] "CHR_HSCHR3_1_CTG2_1"
#> [236] "CHR_HSCHR3_1_CTG3"
#> [237] "CHR_HSCHR3_2_CTG2_1"
#> [238] "CHR_HSCHR3_2_CTG3"
#> [239] "CHR_HSCHR3_3_CTG1"
#> [240] "CHR_HSCHR3_3_CTG3"
#> [241] "CHR_HSCHR3_4_CTG2_1"
#> [242] "CHR_HSCHR3_4_CTG3"
#> [243] "CHR_HSCHR3_5_CTG2_1"
#> [244] "CHR_HSCHR3_5_CTG3"
#> [245] "CHR_HSCHR3_6_CTG3"
#> [246] "CHR_HSCHR3_7_CTG3"
#> [247] "CHR_HSCHR3_8_CTG3"
#> [248] "CHR_HSCHR3_9_CTG3"
#> [249] "CHR_HSCHR4_11_CTG12"
#> [250] "CHR_HSCHR4_1_CTG12"
#> [251] "CHR_HSCHR4_1_CTG4"
#> [252] "CHR_HSCHR4_1_CTG6"
#> [253] "CHR_HSCHR4_1_CTG9"
#> [254] "CHR_HSCHR4_2_CTG12"
#> [255] "CHR_HSCHR4_2_CTG4"
#> [256] "CHR_HSCHR4_3_CTG12"
#> [257] "CHR_HSCHR4_4_CTG12"
#> [258] "CHR_HSCHR4_5_CTG12"
#> [259] "CHR_HSCHR4_6_CTG12"
#> [260] "CHR_HSCHR4_7_CTG12"
#> [261] "CHR_HSCHR4_8_CTG12"
#> [262] "CHR_HSCHR4_9_CTG12"
#> [263] "CHR_HSCHR5_1_CTG1"
#> [264] "CHR_HSCHR5_1_CTG1_1"
#> [265] "CHR_HSCHR5_1_CTG5"
#> [266] "CHR_HSCHR5_2_CTG1"
#> [267] "CHR_HSCHR5_2_CTG1_1"
#> [268] "CHR_HSCHR5_2_CTG5"
#> [269] "CHR_HSCHR5_3_CTG1"
#> [270] "CHR_HSCHR5_3_CTG5"
#> [271] "CHR_HSCHR5_4_CTG1"
#> [272] "CHR_HSCHR5_4_CTG1_1"
#> [273] "CHR_HSCHR5_5_CTG1"
#> [274] "CHR_HSCHR5_6_CTG1"
#> [275] "CHR_HSCHR5_7_CTG1"
#> [276] "CHR_HSCHR6_1_CTG10"
#> [277] "CHR_HSCHR6_1_CTG2"
#> [278] "CHR_HSCHR6_1_CTG3"
#> [279] "CHR_HSCHR6_1_CTG4"
#> [280] "CHR_HSCHR6_1_CTG5"
#> [281] "CHR_HSCHR6_1_CTG6"
#> [282] "CHR_HSCHR6_1_CTG7"
#> [283] "CHR_HSCHR6_1_CTG8"
#> [284] "CHR_HSCHR6_1_CTG9"
#> [285] "CHR_HSCHR6_8_CTG1"
#> [286] "CHR_HSCHR6_MHC_APD_CTG1"
#> [287] "CHR_HSCHR6_MHC_COX_CTG1"
#> [288] "CHR_HSCHR6_MHC_DBB_CTG1"
#> [289] "CHR_HSCHR6_MHC_MANN_CTG1"
#> [290] "CHR_HSCHR6_MHC_MCF_CTG1"
#> [291] "CHR_HSCHR6_MHC_QBL_CTG1"
#> [292] "CHR_HSCHR6_MHC_SSTO_CTG1"
#> [293] "CHR_HSCHR7_1_CTG1"
#> [294] "CHR_HSCHR7_1_CTG4_4"
#> [295] "CHR_HSCHR7_1_CTG6"
#> [296] "CHR_HSCHR7_1_CTG7"
#> [297] "CHR_HSCHR7_2_CTG1"
#> [298] "CHR_HSCHR7_2_CTG4_4"
#> [299] "CHR_HSCHR7_2_CTG6"
#> [300] "CHR_HSCHR7_2_CTG7"
#> [301] "CHR_HSCHR7_3_CTG6"
#> [302] "CHR_HSCHR8_1_CTG1"
#> [303] "CHR_HSCHR8_1_CTG6"
#> [304] "CHR_HSCHR8_1_CTG7"
#> [305] "CHR_HSCHR8_2_CTG1"
#> [306] "CHR_HSCHR8_2_CTG7"
#> [307] "CHR_HSCHR8_3_CTG1"
#> [308] "CHR_HSCHR8_3_CTG7"
#> [309] "CHR_HSCHR8_4_CTG1"
#> [310] "CHR_HSCHR8_4_CTG7"
#> [311] "CHR_HSCHR8_5_CTG1"
#> [312] "CHR_HSCHR8_5_CTG7"
#> [313] "CHR_HSCHR8_6_CTG1"
#> [314] "CHR_HSCHR8_7_CTG1"
#> [315] "CHR_HSCHR8_8_CTG1"
#> [316] "CHR_HSCHR8_9_CTG1"
#> [317] "CHR_HSCHR9_1_CTG1"
#> [318] "CHR_HSCHR9_1_CTG2"
#> [319] "CHR_HSCHR9_1_CTG3"
#> [320] "CHR_HSCHR9_1_CTG4"
#> [321] "CHR_HSCHR9_1_CTG5"
#> [322] "CHR_HSCHR9_1_CTG6"
#> [323] "CHR_HSCHRX_1_CTG3"
#> [324] "CHR_HSCHRX_2_CTG12"
#> [325] "CHR_HSCHRX_2_CTG3"
#> [326] "GL000009.2"
#> [327] "GL000194.1"
#> [328] "GL000195.1"
#> [329] "GL000205.2"
#> [330] "GL000213.1"
#> [331] "GL000216.2"
#> [332] "GL000218.1"
#> [333] "GL000219.1"
#> [334] "GL000220.1"
#> [335] "GL000225.1"
#> [336] "KI270442.1"
#> [337] "KI270711.1"
#> [338] "KI270713.1"
#> [339] "KI270721.1"
#> [340] "KI270726.1"
#> [341] "KI270727.1"
#> [342] "KI270728.1"
#> [343] "KI270731.1"
#> [344] "KI270733.1"
#> [345] "KI270734.1"
#> [346] "KI270744.1"
#> [347] "KI270750.1"
#> [348] "LRG_183"
#> [349] "LRG_187"
#> [350] "LRG_239"
#> [351] "LRG_311"
#> [352] "LRG_721"
#> [353] "LRG_741"
#> [354] "LRG_93"
#> [355] "MT"
#> [356] "X"
#> [357] "Y"
ensembldb::updateEnsDb(x)
#> EnsDb for Ensembl:
#> |Backend: SQLite
#> |Db type: EnsDb
#> |Type of Gene ID: Ensembl Gene ID
#> |Supporting package: ensembldb
#> |Db created by: ensembldb package from Bioconductor
#> |script_version: 0.3.0
#> |Creation time: Thu May 18 16:32:27 2017
#> |ensembl_version: 86
#> |ensembl_host: localhost
#> |Organism: homo_sapiens
#> |taxonomy_id: 9606
#> |genome_build: GRCh38
#> |DBSCHEMAVERSION: 2.0
#> | No. of genes: 63970.
#> | No. of transcripts: 216741.
#> |Protein data available.
ensembldb::genes(x, columns=c("gene_name"),
filter=list(SeqNameFilter("X"), AnnotationFilter::GeneBiotypeFilter("protein_coding")))
#> GRanges object with 841 ranges and 3 metadata columns:
#> seqnames ranges strand | gene_name
#> <Rle> <IRanges> <Rle> | <character>
#> ENSG00000182378 X 276322-303356 + | PLCXD1
#> ENSG00000178605 X 304529-318819 - | GTPBP6
#> ENSG00000167393 X 333963-386955 - | PPP2R3B
#> ENSG00000185960 X 624344-659411 + | SHOX
#> ENSG00000205755 X 1187549-1212750 - | CRLF2
#> ... ... ... ... . ...
#> ENSG00000277745 X 155459415-155460005 - | H2AFB3
#> ENSG00000185973 X 155490115-155669944 - | TMLHE
#> ENSG00000168939 X 155767812-155782459 + | SPRY3
#> ENSG00000124333 X 155881293-155943769 + | VAMP7
#> ENSG00000124334 X 155997581-156010817 + | IL9R
#> gene_id gene_biotype
#> <character> <character>
#> ENSG00000182378 ENSG000001... protein_co...
#> ENSG00000178605 ENSG000001... protein_co...
#> ENSG00000167393 ENSG000001... protein_co...
#> ENSG00000185960 ENSG000001... protein_co...
#> ENSG00000205755 ENSG000002... protein_co...
#> ... ... ...
#> ENSG00000277745 ENSG000002... protein_co...
#> ENSG00000185973 ENSG000001... protein_co...
#> ENSG00000168939 ENSG000001... protein_co...
#> ENSG00000124333 ENSG000001... protein_co...
#> ENSG00000124334 ENSG000001... protein_co...
#> -------
#> seqinfo: 1 sequence from GRCh38 genome
ensembldb ::transcripts(x, columns=ensembldb::listColumns(x, "tx"),
filter = AnnotationFilter::AnnotationFilterList(), order.type = "asc", return.type = "GRanges")
#> GRanges object with 216741 ranges and 6 metadata columns:
#> seqnames ranges strand | tx_id
#> <Rle> <IRanges> <Rle> | <character>
#> ENST00000456328 1 11869-14409 + | ENST000004...
#> ENST00000450305 1 12010-13670 + | ENST000004...
#> ENST00000488147 1 14404-29570 - | ENST000004...
#> ENST00000619216 1 17369-17436 - | ENST000006...
#> ENST00000473358 1 29554-31097 + | ENST000004...
#> ... ... ... ... . ...
#> ENST00000420810 Y 26549425-26549743 + | ENST000004...
#> ENST00000456738 Y 26586642-26591601 - | ENST000004...
#> ENST00000435945 Y 26594851-26634652 - | ENST000004...
#> ENST00000435741 Y 26626520-26627159 - | ENST000004...
#> ENST00000431853 Y 56855244-56855488 + | ENST000004...
#> tx_biotype tx_cds_seq_start tx_cds_seq_end gene_id
#> <character> <integer> <integer> <character>
#> ENST00000456328 processed_... <NA> <NA> ENSG000002...
#> ENST00000450305 transcribe... <NA> <NA> ENSG000002...
#> ENST00000488147 unprocesse... <NA> <NA> ENSG000002...
#> ENST00000619216 miRNA <NA> <NA> ENSG000002...
#> ENST00000473358 lincRNA <NA> <NA> ENSG000002...
#> ... ... ... ... ...
#> ENST00000420810 processed_... <NA> <NA> ENSG000002...
#> ENST00000456738 unprocesse... <NA> <NA> ENSG000002...
#> ENST00000435945 unprocesse... <NA> <NA> ENSG000002...
#> ENST00000435741 processed_... <NA> <NA> ENSG000002...
#> ENST00000431853 processed_... <NA> <NA> ENSG000002...
#> tx_name
#> <character>
#> ENST00000456328 ENST000004...
#> ENST00000450305 ENST000004...
#> ENST00000488147 ENST000004...
#> ENST00000619216 ENST000006...
#> ENST00000473358 ENST000004...
#> ... ...
#> ENST00000420810 ENST000004...
#> ENST00000456738 ENST000004...
#> ENST00000435945 ENST000004...
#> ENST00000435741 ENST000004...
#> ENST00000431853 ENST000004...
#> -------
#> seqinfo: 357 sequences (1 circular) from GRCh38 genome
#txdbEnsemblGRCh38 <- GenomicFeatures::makeTxDbFromEnsembl(organism="Homo sapiens", release=98)
#txdb <- as.list(txdbEnsemblGRCh38)
#lapply(txdb,head)11 Spectrum data analysis
This section collects notes on peptide/protein analysis, especially with respect to spectrum data.
11.1 Peptide sequence
Here is an example for PROC_HUMAN, which is handled by the Biostrings package,
fasta_file_path <- 'https://rest.uniprot.org/uniprotkb/P04070.fasta'
fasta_sequences <- Biostrings::readAAStringSet(fasta_file_path, format = "fasta")
AA_sequence <- fasta_sequences[[1]]
cat("Sequence:", toString(AA_sequence), "\n")
iso_442688365 <- 'TDGEGALSEPSATVTIEELAAPPPPVLMHHGESSQVLHPGNK'
match_position <- regexpr(iso_442688365, AA_sequence)
match_position
mp <- matchPattern(iso_442688365,AA_sequence)
mp
load(system.file("tests","PROC.rda",package="pQTLtools"))
pQTLtools::peptideAssociationPlot(protein,cistrans)
#> Joining with `by = join_by(Modified.Peptide.Sequence)`
Figure 11.1: peptide association plot
11.2 Spectrum data analysis
11.2.1 Setup
The .raw files can be handled by rawrr package nevertheless it requires necessary files,
library(rawrr)
if (isFALSE(rawrr::.checkDllInMonoPath())){
rawrr::installRawFileReaderDLLs()
}
if (isFALSE(file.exists(rawrr:::.rawrrAssembly()))){
rawrr::installRawrrExe()
}11.2.2 List of.raw files
Based on a real project, the following is an example of listing/generating multiple .raw from .zip files
# ZWK .raw data
spectra_ZWK <- "~/Caprion/pre_qc_data/spectral_library_ZWK"
raw_files <- list.files(spectra_ZWK, pattern = "\\.raw$", full.names = TRUE)
## collectively
suppressMessages(library(MsBackendRawFileReader))
ZWK <- Spectra::backendInitialize(MsBackendRawFileReader::MsBackendRawFileReader(),
files = raw_files)
class(ZWK)
methods(class=class(ZWK))
Spectra(ZWK)
spectraData(ZWK)
ZWK
ZWKvars <- ZWK |> Spectra::spectraVariables()
ZWKdata <- ZWK |> Spectra::spectraData()
dim(ZWKdata)
# rows with >=1 non-NA value in the columns with prefix "precursor"
precursor <- apply(ZWKdata[grep("precursor",ZWKvars)], 1, function(x) any(!is.na(x)))
ZWKdata_filtered <- ZWKdata[precursor, ]
save(ZWK,file="~/Caprion/analysis/work/ZWK.rda")
# ZYQ/UDP
library(utils)
spectra <- "~/Caprion/pre_qc_data/spectra"
zip_files <- dir(spectra, recursive = TRUE, full.names=TRUE)
work_dir <- "~/Caprion/analysis/work"
for (zip_file in zip_files) unzip(zip_file, exdir=work_dir)
ZYQ_UDP <- Spectra::backendInitialize(MsBackendRawFileReader::MsBackendRawFileReader(),
files = dir(work_dir,patt="raw",full.names=TRUE))
class(ZYQ_UDP)
ZYQ_UDP
ZYQ_UDP |> Spectra::spectraVariables()
save(ZYQ_UDP,file="~/Caprion/analysis/work/ZYQ_UDP.rda")11.2.3 Usage
Various facilities are shown below.
# various files
d <- "/rds/project/rds-zuZwCZMsS0w/Caprion_proteomics/analysis/crux"
f <- file.path(d,"szwk901104i19801xms1.mzML")
x <- file.path(d,"szwk901104i19801xms1.mzXML")
g <- file.path(d,"szwk901104i19801xms1.mgf")
r <- file.path(d,"szwk901104i19801xms1.rda")
z <- file.path(d,"szwk901104i19801xms1.mzML.gz")
# mzML
mz <- mzR::openMSfile(f)
header_info <- mzR::header(mz)
table(header_info$msLevel)
peak_data <- mzR::peaks(mz)
spec <- mzR::spectra(mz)
class(spec)
length(spec)
lapply(spec,head,3)
methods(class="mzRpwiz")
mzR::close(mz)
mz <- mzR::openMSfile(z, backend = "pwiz")
mz
nChrom(mz)
head(tic(mz))
head(chromatogram(mz, 1L)) ## same as tic(x)
str(chromatogram(mz))
head(peaks(mz, scan=4))
# MSnbase
mzXML <- MSnbase::readMSData(x)
mgf <- MSnbase::readMgfData(g)
save(mzXML,mgf,file=r)
MSnbase::extractSpectraData(mzXML)
MSnbase::hasSpectra(z)
MSnbase::hasChromatograms(z)
MSnbase::plot2d(mzXML,z="peaks.count")
MSnbase::plotDensity(mzXML,z="precursor.mz")
MSnbase::extractSpectraData(mgf)
methods(class="MSpectra")
MSnbase::mz(mgf)
MSnbase::intensity(mgf)
MSnbase::rtime(mgf)
MSnbase::precursorMz(mgf)
MSnbase::precursorCharge(mgf)
MSnbase::precScanNum(mgf)
MSnbase::precursorIntensity(mgf)
MSnbase::acquisitionNum(mgf)
MSnbase::scanIndex(mgf)
MSnbase::peaksCount(mgf)
MSnbase::msLevel(mgf)
MSnbase::tic(mgf)
MSnbase::ionCount(mgf)
MSnbase::collisionEnergy(mgf)
MSnbase::fromFile(mgf)
MSnbase::polarity(mgf)
MSnbase::smoothed(mgf)
MSnbase::centroided(mgf)
MSnbase::isCentroided(mgf)
MSnbase::writeMgfData(mgf, con = "spectra.mgf", COM = NULL, TITLE = NULL)
MSnbase::removePeaks(mgf, t, msLevel., ...)
MSnbase::filterMsLevel(mgf, msLevel=2)
MSnbase::as.ExpressionSet(mgf)
# This turned to be really slow!
sp_list <- lapply(seq_along(mgf), function(i) {
intensity_i <- MSnbase::intensity(mgf)[[i]]
mz_i <- MSnbase::mz(mgf)[[i]]
centroided_i <- MSnbase::centroided(mgf)[[i]]
return(new("Spectrum1", intensity = intensity_i, mz = mz_i, centroided = centroided_i))
})
sp1 <- do.call(rbind, sp_list)
# only the first one is more manageable
sp1 <- new("Spectrum1",intensity=MSnbase::intensity(mgf)[[1]],mz=MSnbase::mz(mgf)[[1]],centroided=MSnbase::centroided(mgf)[[1]])
sp2 <- MSnbase::pickPeaks(sp1)
MSnbase::intensity(sp2)
plot(MSnbase::mz(sp1),MSnbase::intensity(sp1),type="h")
## Without m/z refinement
points(MSnbase::mz(sp2), MSnbase::intensity(sp2), col = "darkgrey")
## Using k = 1, closest signals
sp3 <- MSnbase::pickPeaks(sp1, refineMz = "kNeighbors", k = 1)
points(MSnbase::mz(sp3), MSnbase::intensity(sp3), col = "green", type = "h")
## Using descendPeak requiring at least 50% or the centroid's intensity
sp4 <- MSnbase::pickPeaks(sp1, refineMz = "descendPeak", signalPercentage = 50)
points(MSnbase::mz(sp4), MSnbase::intensity(sp4), col = "red", type = "h")
# CAMERA
xs <- CAMERA::xcmsSet(f, method="centWave", ppm=30, peakwidth=c(5,10))
an <- CAMERA::xsAnnotate(xs)
an <- CAMERA::groupFWHM(an)
#For one group
peaklist <- CAMERA::getpspectra(an, 1)
#For two groups
peaklist <- CAMERA::getpspectra(an, c(1,2))
# Spectra
suppressMessages(library(Spectra))
sp <- Spectra::Spectra(z)
head(sp)
table(sp$msLevel)
d <- Spectra::computeMzDeltas(sp[1:1000])
Spectra::plotMzDelta(d)
# protViz
protViz::fragmentIon("TFVLNFIK")
esd <- MSnbase::extractSpectraData(mgf)
op <- par(mfrow=c(2,1))
ms <- function(i) with(esd[i,],list(title=TITLE,rtinseconds=RTINSECONDS,pepmass=PEPMASS,charge=CHARGE,
mZ=MSnbase::mz(mgf[[i]]),intensity=MSnbase::intensity(mgf[[i]])))
protViz::peakplot("TAFDEAIAELDTLNEESYK", ms(1))
protViz::peakplot("TAFDEAIAELDTLSEESYK", ms(2))
par(op)
load("~/Caprion/pilot/ZWK.rda")
peptides <- subset(mapping_ZWK,Protein=="PROC_HUMAN")[["Modified.Peptide.Sequence"]] |> unique()
pim <- protViz::parentIonMass(peptides)
fi <- protViz::fragmentIon(peptides)
df <- as.data.frame(fi)
op <- par(mfrow=c(3,1))
for (i in 1:length(peptides)){
plot(0, 0,
xlab='m/Z',
ylab='',
xlim=range(c(fi[[i]]$b,fi[[i]]$y)),
ylim=c(0,1),
type='n',
axes=FALSE,
sub=paste(peptides[i], "/", pim[i], "Da"));
box()
axis(1, fi[[i]]$b, round(fi[[i]]$b,1), las=2)
axis(1, fi[[i]]$y, round(fi[[i]]$y,1), las=2)
pepSeq<-strsplit(peptides[i], "")
axis(3,fi[[i]]$b, paste("b", row.names(fi[[i]]),sep=''),las=2)
axis(3,fi[[i]]$y, paste("y", row.names(fi[[i]]),sep=''),las=2)
text(fi[[i]]$b, rep(0.3, nchar(peptides[i])),
pepSeq[[1]],pos=3,cex=4, lwd=4, col="#aaaaaaaa")
abline(v=fi[[i]]$b, col='red')
abline(v=fi[[i]]$y, col='blue',lwd=2)
}
par(op)
# MSstats
head(SRMRawData)
QuantData <- MSstats::dataProcess(SRMRawData, use_log_file = FALSE)
quant <- MSstats::dataProcess(SRMRawData,
normalization = "equalizeMedians",
summaryMethod = "TMP",
censoredInt = "NA",
MBimpute = TRUE,
maxQuantileforCensored = 0.999,
logTrans=2,
use_log_file=FALSE,
numberOfCores=5)
names(quant)MSstats3 takes output from other data-processing pipelines.
13 Bioconductor/CRAN packages
| Package | Description |
|---|---|
| Bioconductor | |
| AnnotationDbi | AnnotationDb objects and their progeny, methods etc. |
| Biobase | Base functions for Bioconductor |
| Biostrings | Efficient manipulation of biological strings |
| CAMERA | Collection of annotation related methods |
| clusterProfiler | Functional profiles for genes and gene clusters |
| ComplexHeatmap | Make complex heatmaps |
| DESSeq2 | Differential gene expression analysis based on the negative binomial distribution |
| edgeR | Empirical analysis of digital gene expression |
| EnsDb.Hsapiens.v86 | Exposes an annotation databases generated from Ensembl |
| ensembldb | Retrieve annotation data from an Ensembl based package |
| FlowSorted.DLPFC.450k | Illumina HumanMethylation data on sorted frontal cortex cell populations |
| graphite | GRAPH Interaction from pathway topological environment |
| IlluminaHumanMethylation450kmanifest | Annotation for Illumina’s 450k methylation arrays |
| INSPEcT | Quantification of the intronic and exonic gene features and the post-transcriptional regulation analysis |
| MSnbase | Base Functions and Classes for Mass Spectrometry and Proteomics |
| MSstats | Protein Significance Analysis in DDA, SRM and DIA Proteomics |
| mzR | parser for netCDF, mzXML and mzML and mzIdentML files |
| org.Hs.eg.db | Conversion of Entrez ID – gene symbols |
| OUTRIDER | OUTlier in RNA-Seq fInDER |
| Pi | Priority index, leveraging genetic evidence to prioritise drug targets at the gene and pathway level |
| quantro | A test for when to use quantile normalisation |
| rawrr | Direct Access to Orbitrap Data and Beyond |
| recount3 | Interface to uniformly processed RNA-seq data |
| Rgraphiz | Interfaces R with the AT&T graphviz library for plotting R graph objects from the graph package |
| Spectra | Spectra Infrastructure for Mass Spectrometry |
| sva | Surrogate Variable Analysis |
| TxDb.Hsapiens.UCSC.hg38.knownGene | Annotation of the human genome |
| CRAN | |
| doParallel | Foreach Parallel Adaptor for the ‘parallel’ Package |
| GeneNet | Modeling and Inferring Gene Networks |
| ggplot2 | Data Visualisations Using the grammar of graphics |
| heatmaply | Interactive Cluster Heat Maps Using plotly and ggplot2
|
| pheatmap | results visualisation |
| plyr | Splitting, applying and combining data |
| protViz | Foreach Parallel Adaptor for the ‘parallel’ Package |
| RColorBrewer | ColorBrewer Palettes |
| WGCNA | Weighted correlation network analysis |